Figure 2.
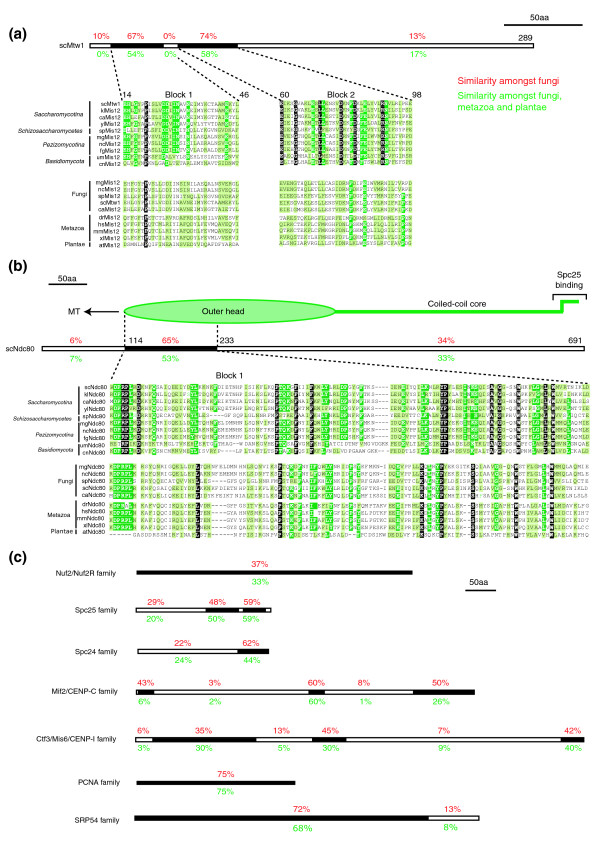
Sequence similarity between kinetochore proteins is restricted to short stretches between orthologs. Multiple sequence alignments of the (a) Mis12Mtw1 and (b) Ndc80Hec1 families. Schematic drawing above the alignment indicate the length of the S. cerevisiae proteins and the percentages denote the degree of similarity of successive sequence blocks (black boxes) within fungi (red letters) or fungi, metazoa and plantae (green letters). The schematic drawing above the Ndc80 multiple sequence alignment also indicates the relative position of the globular and coiled-coil domain of Ndc80, as determined by electron-microscopy [32,33]. White letters on black denote identical residues, white letters on green, identical residues in ≥ 80% of the organisms and black letters on green, similar residues in ≥ 80% of the organisms. (c) Schematic drawings indicating the percentage similarity of successive sequence blocks (black boxes) within fungi (red letters) or fungi, metazoa and plantae (green letters) based on multiple sequence alignments of the Nuf2, Spc25, Spc24. CENP-CMif2 and Mis6Ctf3/CENP-I, PCNA and SRP54 protein families
