Figure 3.
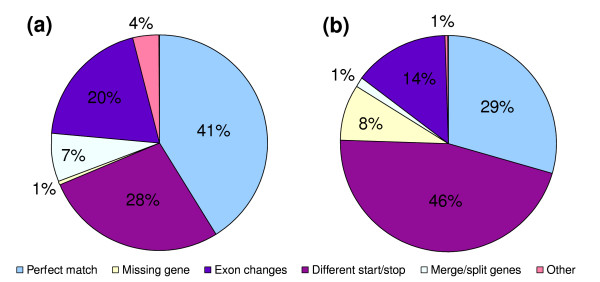
Manual comparison of ReAnoCDS05 and Ensembl based on a set of full-length cDNA sequences. The charts show the analysis of all cDNAs in the dataset mapped to the X-chromosome (n = 800), corresponding to 156 cDNA loci, and their conceptual translation products in relation to CDSs predicted by (a) ReAnoCDS05 and (b) Ensembl. Categories of comparison indicated in the legend are: perfect match, proportion of cDNA sequences with translation products that display exact match to predicted peptide sequence of annotation CDS; missing gene, cDNAs not represented by a corresponding annotation CDS; exon changes, cDNAs for which the corresponding annotation CDSs display extra exons, missing exons, and/or exon boundary changes; different start/stop, cDNA loci for which annotation CDSs display different predicted translation initiation and/or termination; merge/split genes, cDNA loci that overlap multiple annotation CDSs, or vice versa; other, including multiple low-frequency cases.
