Figure 6.
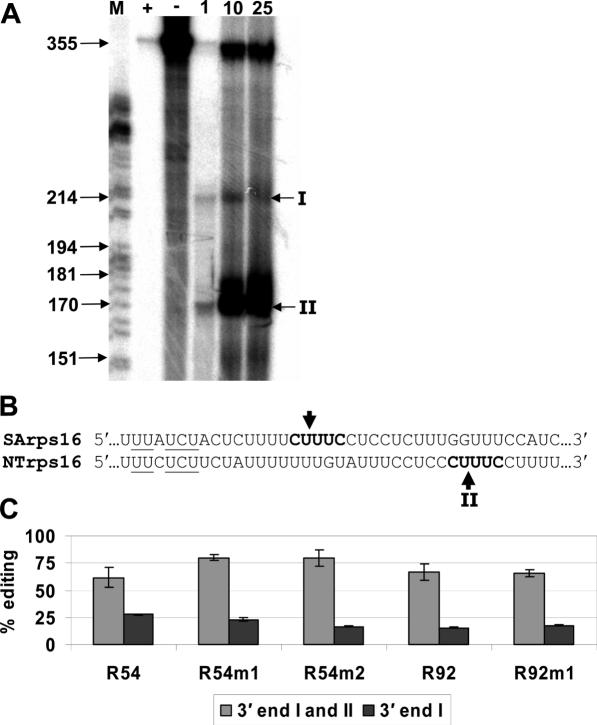
The 3′ ends of transgenic transcripts were determined using S1 nuclease protection mapping. (A) The 355 nt antisense probe was mixed with S1 nuclease (lane +), without S1 nuclease (lane −), and 1, 10 and 25 μg (lanes 1, 10, 25) of total RNA from leaves of transplastomic shoots. Two 3′ ends were observed (I and II). Lane M contains a DNA sequencing reaction, which served as a molecular weight standard. (B) The 3′-UTR sequences from rps16 of Sinapsis alba (SArps16) and N.tobacum (NTrps16) were aligned. Arrows indicate mapped cleavage sites and underlined characters represent a 7mer protein-binding region from Nickelsen and Link (27). (C) The percentage of edited transgenic transcripts calculated by poisoned primer extension reactions with either 3′ end I or both 3′ ends. Error bars represent 1 SD from the mean.