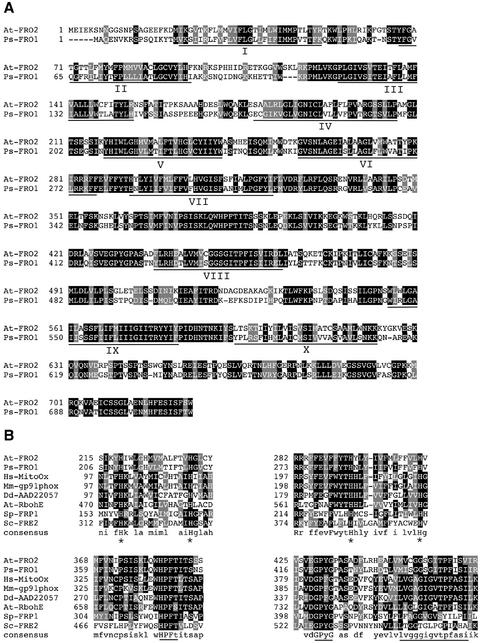
Figure 1.
Characterization of the FRO1 amino acid sequence. A, Alignment of Arabidopsis FRO2 (At-FRO2) and pea FRO1 (Ps-FRO1). Positions of amino acid identity are shaded in black, and similar residues are shaded in gray. The locations of the 10 potential transmembrane domains are underlined and numbered I-X. B, Conservation of specific motifs within FRO1 and related oxidoreductases. FRO1 and FRO2 sequences are aligned with human mitogenic oxidase (Hs-MitoOx, accession no. AF127763), mouse gp91phox (Mm-gp91phox, accession no. U43384), Dictyostelium discoideum superoxide-generating NADPH oxidase heavy chain (Dd-AAD22057, accession no. AF123275), Arabidopsis RbohE (At-RbohE, accession no. AF055356), S. pombe FRP1 (Sp-FRP1, accession no. L07749), and S. cerevisiae FRE2 (Sc-FRE2, accession no. Z28220). Conserved histidines involved in heme binding are indicated with asterisks, and the conserved FAD-binding motif (HPFT), the NAD-binding motif (GPyG), and the oxidoreductase signature sequence are underlined.