Abstract
In plants, fatty acid and complex lipid synthesis requires the correct spatial and temporal activity of many gene products. Quantitative northern analysis showed that mRNA for the biotin carboxylase subunit of heteromeric acetyl-coenzyme A carboxylase, fatty acid synthase components (3-oxoacyl-acyl carrier protein [ACP] reductase, enoyl-ACP reductase, and acyl-ACP thioesterase), and stearoyl-ACP desaturase accumulate in a coordinate manner during Brassica napus embryogenesis. The mRNAs were present in a constant molar stoichiometric ratio. Transcript abundance of mRNAs for the catalytic proteins was found to be similar, whereas the number of ACP transcripts was approximately 7-fold higher. The peak of mRNA accumulation of all products was between 20 and 29 d after flowering; by 42 d after flowering, the steady-state levels of all transcripts fell to about 5% of their peak levels, which suggests that the mRNAs have similar stability and kinetics of synthesis. Biotin carboxylase was found to accumulate to a maximum of 59 fmol mg−1 total RNA in embryos, which is in general agreement with the value of 170 fmol mg−1 determined for Arabidopsis siliques (J.S. Ke, T.N. Wen, B.J. Nikolau, E.S. Wurtele [2000] Plant Physiol 122: 1057–1071). Embryos accumulated between 3- and 15-fold more transcripts per unit total RNA than young leaf tissue; the lower quantity of leaf 3-oxoacyl-ACP reductase mRNA was confirmed by reverse transcriptase-polymerase chain reaction. This is in conflict with analysis of B. napus transcripts using an Arabidopsis microarray (T. Girke, J. Todd, S. Ruuska, J. White, C. Benning, J. Ohlrogge [2000] Plant Physiol 124: 1570–1581) where similar leaf to seed levels of fatty acid synthase component mRNAs were reported.
Fatty acids are synthesized by a common biochemical pathway in all organisms. In plants, de novo synthesis takes place in plastids, using two enzyme systems: acetyl-CoA carboxylase (ACCase) and fatty acid synthase (FAS). The type II FAS, of plants, is composed of separate soluble enzymes that each carry out a single enzymatic reaction (Caughey and Kekwick, 1982; Hoj and Mikkelsen, 1982; Shimakata and Stumpf, 1982) with the growing acyl chains attached to acyl carrier protein (ACP). This is in contrast to the arrangement in animals and yeast where the enzyme activities and ACP are located on one or two multifunctional polypeptides (type I FAS).
Acetyl-CoA is carboxylated to malonyl-CoA in plastids of non-graminaceous plants by a heteromeric ACCase, which is encoded by four subunits (Sasaki et al., 1993, 1995). The α-carboxyltransferase (α-CT) polypeptide is encoded in the plastid genome (Sasaki et al., 1993), whereas the β-carboxyltransferase (β-CT), biotin carboxyl carrier protein (BCCP) and biotin carboxylase (BC) subunits are nuclear encoded. Malonyl-CoA undergoes a thiol-exchange reaction carried out by malonyl-CoA:ACP transacylase (MCAT) to form malonyl-ACP, before condensation with acetyl-CoA. The initial condensation reaction, catalyzed by ketoacyl-ACP synthase (KAS) III, is unique in that malonyl-ACP reacts with acetyl-CoA. Subsequent condensations add two carbon units from malonyl-ACP to the saturated acyl-ACP chain by the action of KAS isoforms I and II. The 3-oxo group is sequentially reduced to a methylene group by the action of 3-oxoacyl-ACP reductase (KR), hydroxyacyl-ACP dehydrase (DH), and enoyl-ACP reductase (ENR) via hydroxyl and enoyl intermediates, before a further condensation reaction takes place.
When the acyl chain is 16 or 18 carbons long, several possible reactions occur in plastids. The saturated acyl-ACP may have a double bond introduced, between carbons 9 and 10, by acyl-ACP desaturase (DES). As an alternative, the acyl chain may be transferred to glycerol-3-phosphate by glycerol-3-phosphate acyltransferase (G3PAT) in the first step of plastid glycerolipid biosynthesis or may be acted upon by acyl-ACP thioesterase (TE), which removes the ACP group in preparation for export of the acyl chain from the plastid. These reactions are carried out by soluble enzymes; membrane-associated enzymes in the plastid or endoplasmic reticulum carry out further steps in the synthesis of complex lipids, for membranes, signaling, and storage. In seeds of many plants, the acyl chain is elongated in the cytoplasm before incorporation into storage triglycerides. The malonyl units used in these condensation reactions are formed by the action of a homomeric ACCase.
Plants face a considerable challenge in matching the tissue-specific and temporal demands for acyl chains and complex lipids with their supply. Regulation of synthesis must be closely controlled at one or more levels, either transcriptionally, at the point of translation or post-translationally. Recent evidence suggests that the β-CT domain of pea chloroplast ACCase may be regulated by phosphorylation (Savage and Ohlrogge, 1999). However, a significant regulatory mechanism for FAS and lipid biosynthetic genes appears to be at the level of transcription (Slocombe et al., 1992; Elborough et al., 1994; Fawcett et al., 1994).
The demand for tissue-specific and temporal increases in FAS components is satisfied in different ways. Multigene families encode some FAS and lipid biosynthetic components. For example, ACP is encoded by at lease five genes in Arabidopsis; both constitutive and tissue-specific patterns of expression have been observed in Arabidopsis (Hlousek-Radojcic et al., 1992; Baerson et al., 1994) and Brassica napus (de Silva et al., 1990). ENR is conversely encoded by a single gene in Arabidopsis that is up-regulated in embryos during increased demand for fatty acids supply (de Boer et al., 1998). Promoter analysis of the Arabidopsis ENR gene revealed that high-level seed expression was maintained in a (−)47 promoter deletion, indicating that all of the cis elements necessary were present on this small region of DNA (de Boer et al., 1999). In the related crop plant B. napus, similar ENR expression patterns were observed; here, ENR is encoded by four genes, each of which is expressed in leaf tissue and also in embryos at higher levels (Fawcett et al., 1994).
To ensure the availability of the necessary gene products for lipid biosynthesis, it is likely that the genes are coordinately expressed. It was recently demonstrated that the components of chloroplastic ACCase (BC, BCCP, α-CT, and β-CT) maintain a constant molar stoichiometric mRNA ratio in siliques throughout development. In addition, these ratios are also maintained in other tissue, such as young leaves and flowers, which are rapidly synthesizing lipids (Ke et al., 2000). To use optimally the supply of malonyl-CoA, other genes involved in fatty acid and lipid metabolism may also be expressed with similar tissue and temporal patterns.
In this report, we determine the temporal pattern of ACP, KR, BC, TE, and ENR mRNA accumulation during B. napus embryogenesis. These studies establish that FAS component mRNAs accumulate at a constant ratio but at different absolute levels. The central FAS components accumulate at similar low levels, whereas those gene products that act upon mature acyl-ACP substrates accumulate at higher levels. The highest level of mRNA accumulation observed was that of ACP. Coordinate expression of FAS components, lipid biosynthetic components, and genes that encode heteromeric ACCase suggests that the steady-state levels of mRNA is an important factor in developmental regulation of enzyme activities.
RESULTS
Hybridization of Probes to Lipid Biosynthetic Gene Family Members
One aim of these experiments was to determine, wherever possible, the mRNA abundances for all members of the gene families investigated. To investigate whether this was possible, using single gene family members as probes, a database search was undertaken to determine the cDNA sequence similarity of B. napus lipid biosynthetic gene family members deposited in GenBank.
Complete data sets were available for the cDNA sequences encoding all ENR and KR family members from B. napus. The mature protein coding regions of the ENR sequences (AJ243087–90) share greater than 89% identity. Similar high homologies exist between the four cDNAs of the KR gene family (AJ243083–6), with the coding regions being greater than 90% identical at the nucleotide level.
Data for the other B. napus lipid biosynthetic gene families was not complete in GenBank; there were two sequences for both DES and TE and single sequences for MCAT, BC, and ACCaseI. Comparison of these sequences with those from the related crucifer Arabidopsis revealed greater than 85% sequence identity over the region of the probes used. Therefore, a washing stringency of 79% identity was chosen to allow hybridization to all mRNAs in the ENR, KR, BC, MCAT, SD, TE, and ACCaseI gene families.
One known exception to the high sequence conservation seen in lipid biosynthetic gene families is that of ACP. Analysis of the mature coding regions of Arabidopsis ACP-1, ACP-2, and ACP-3 genes shows that they are more divergent, sharing a minimum of 69% identity (Hlousek-Radojcic et al., 1992). The embryo-expressed ACP probe used in this study shares greater than 85% identity to known B. napus cDNA sequences, many of which were cloned from an embryo cDNA library. These sequences represent six unique genes (Safford et al., 1988) from the ACP family of approximately 35 genes present per haploid genome (de Silva et al., 1990). Therefore, it is possible that the ACP probe would not hybridize to all members of the B. napus ACP gene family; it would, however, hybridize to all known embryo-expressed sequences.
Accumulation of FAS mRNAs
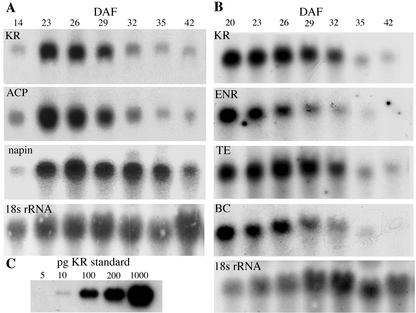
An important means of regulation of FAS activity is thought to be at the level of transcription (Elborough et al., 1994; Fawcett et al., 1994; Slocombe et al., 1994). Members of the fatty acid and lipid biosynthetic pathway were analyzed for coordinate accumulation of their mRNAs (Fig. 1). The mass of each specific sense mRNA was determined by comparison with hybridization to in vitro generated antisense mRNA standards of known concentration. These analyses were carried out using RNA gel blots; each blot contained standard amounts of KR mRNA and either ACP or ENR, TE, and BC sequences. The RNA blots indicated apparent coordination in the steady-state levels of the five components tested (Fig. 1). Each of the components exhibited high levels of mRNA accumulation in the 20 to 29 d after flowering (DAF) period, which was followed by dramatic reductions at 32 DAF. The increased demand for fatty acid synthesis during embryogenesis was, therefore, initially met by increased accumulation of FAS component mRNAs.
Figure 1.
Steady-state mRNA levels during embryogenesis. A, The profile for KR and ACP; B, the profile for KR, ENR, TE, and BC. C, Hybridization to a dilution series of antisense KR mRNA (5–1,000 pg) that was loaded into a second series of wells before electrophoresis was completed. Results were normalized after reprobing with an 18S rRNA sequence. The seed storage protein napin gene was used to probe blot A and showed a different temporal steady-state expression pattern.
The temporal expression pattern of FAS genes markedly differed from that of the gene for the storage protein napin. The napin protein starts to accumulate approximately halfway through embryo development, when storage oil synthesis is occurring at a maximum rate, and continues to be synthesized until the seed starts to desiccate (Murphy and Cummins, 1989). The disparity in steady-state levels of FAS and napin mRNAs can be explained by the fact that napin encodes for a storage protein, whereas FAS mRNAs encode catalytic proteins that require smaller numbers of translated products.
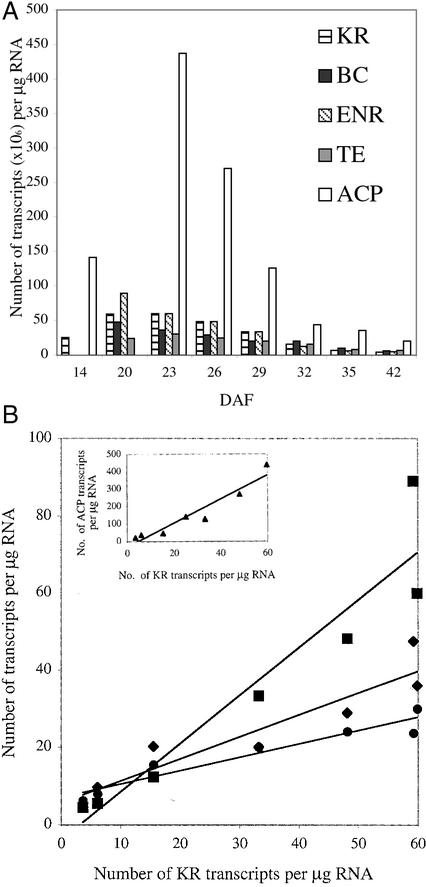
After adjusting for RNA loading by comparing the abundance of 18S rRNA transcript in each lane, the mass of FAS mRNAs detected and their transcript sizes were used to calculate the number of transcripts per microgram of total RNA (Fig. 2A). The transcript abundance of mRNAs for each of the catalytic proteins (BC, KR, ENR, and TE) was found to be similar, whereas the number of ACP transcripts was approximately 7-fold higher than that of KR. In the 42 DAF sample, the steady-state levels of all transcripts fell to about 5% of the peak level. The decrease in transcript levels occurred in a coordinate fashion, which suggests that the mRNAs have similar stability and rates of synthesis.
Figure 2.
Coordinate changes in accumulation of mRNAs. A, The concentration of KR, ACP, ENR, TE, and BC mRNAs in each sample was determined by comparing with hybridization intensity of RNA standards and was used to calculate the number of transcripts. B, The number of KR transcripts at each time point in embryogenesis was plotted against the number of transcripts for ENR (▪), BC (♦), TE (●), and ACP (▴). Values are ×106 per microgram of total RNA. Analysis of the data indicates linear relationship to the number of KR transcripts with r2 values of 0.90 (ENR), 0.88 (BC), 0.92 (TE), and 0.92 (ACP).
FAS Components Exhibit Constant mRNA Ratios
The coordinate nature of FAS mRNA accumulation was analyzed by plotting the number of transcripts for BC, ENR, TE, and ACP at each time point throughout embryogenesis against the number of KR transcripts (Fig. 2B). These plots revealed a linear relationship between the numbers of lipid synthesis components. The ratio between the mRNAs was calculated to be KR:BC:ENR:TE:ACP = 1.0:0.6:1.2:0.4:6.9; the regression coefficients for these data were greater than 0.88 in each case. This indicates that mRNAs involved in lipid biosynthesis accumulate at a constant molar ratio throughout embryo development.
Quantitative RNA blots were also used to determine the amount of transcripts present in B. napus embryos. Repeat blots using RNA standards generated from different reverse transcriptase (RT) reactions give a range for the maximum accumulation of KR mRNA of between 40 and 99 fmol mg−1 total RNA. Data were generally consistent with the higher value, which was used to calculate the number of KR transcripts.
The maximum concentration of BC mRNA was 59 fmol mg−1 total RNA. This value is in general agreement with that for Arabidopsis silique, which has been reported to contain a maximum of 170 fmol BC mRNA mg−1 total RNA (Ke et al., 2000).
DNA Array Analysis of FAS and Lipid Biosynthesis Genes
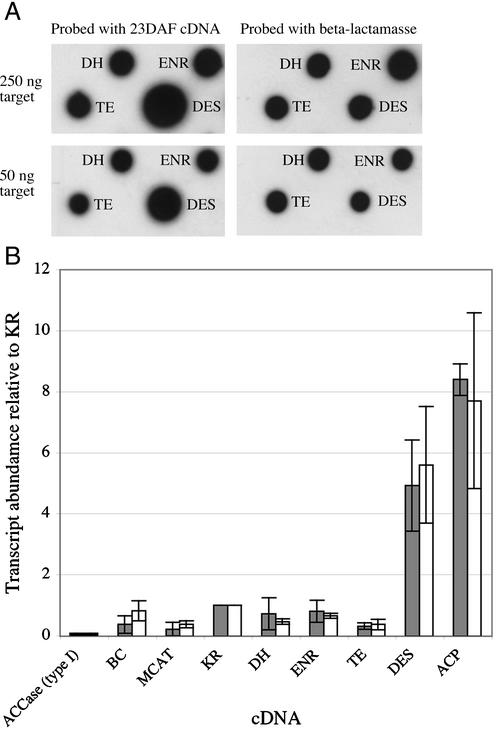
To screen a larger number of lipid biosynthetic components at one time and to verify the RNA-blot data, we employed a DNA filter array containing nine cDNA sequences (present in the plasmid vector pBluescript SK+) involved in lipid biosynthesis and a cDNA encoding napin. Duplicate nylon membranes, each containing 50 and 250 ng of target cDNA clones were probed with first-strand cDNA from 23 DAF embryos. Figure 3A shows a portion of the filter array. It is clear that the steady-state level of DES is much higher than that of TE, and the level of hybridization of ENR was qualitatively similar to that of DH. The filters were subsequently probed with a labeled β-lactamase sequence (Fig. 3A). Because this was present on each plasmid in a 1:1 ratio with the cDNA, the results were used to normalize for target loading. Transcript abundances of the genes, relative to the level of KR are shown in Figure 3B; the two different levels of target cDNA gave similar results.
Figure 3.
Filter-array analysis of FAS and lipid biosynthetic genes. A, Sections of the filter showing high expression of DES compared with TE. The filters were hybridized with first-strand cDNA from 23 DAF embryo mRNA and β-lactamase, which was used as a loading control. Analysis was carried out on duplicate blots containing 50 and 250 ng of each target sequence. B, Transcript abundance of components of lipid biosynthesis, relative to KR = 1.0. Filled bars represent 50 ng and white bars 250 ng of each target sequence.
There was good correlation between the array analysis and results from quantitative northern blots (Table I), for example, the ACP:KR ratio from the array was 8.1:1 and the northern analysis gave a result of 6.9:1. The FAS components KR, DH, and ENR that are involved in the reduction of oxo-groups before further condensation reactions, and the reactions that supply malonyl-ACP (BC and MCAT) have similar steady-state levels of mRNA using either method (Table I). Two transcripts were seen to accumulate at a higher level: ACP, which exhibited the highest steady-state level, and DES, whose gene product carries out reactions on mature acyl-ACPs.
Table I.
Comparison of experimentally determined transcript abundance with corresponding entries in dbESTs
| Gene | Macro-Array (23 DAF) | Quantitative Northern (23 DAF) | Plant dbEST Entriesa | Seed EST Datab |
|---|---|---|---|---|
| ACCaseI | 0.1 | 0.6 (12) | ||
| BC | 0.6 | 0.6 | 0.8 (16) | 0.125 (2) |
| MCAT | 0.3 | 0.5 (10) | ||
| KR | 1.0 | 1.0 | 1.0 (20) | 1.0 (16) |
| DH | 0.6 | 0.4 (8) | 0 (0) | |
| ENR | 0.7 | 1.2 | 1.15 (23) | 0 (0) |
| TE | 0.3 | 0.4 | 0.35 (7) | |
| DES | 5.3 | 2.65 (53) | 1.06 (17) | |
| ACP | 8.1 | 6.9 | 4.45 (89) | |
| Napin | 60 |
Each transcript is represented as a ratio to the abundance of KR. The numbers of entries analyzed from plant dbESTs are given in parentheses.
From Mekhedov et al. (2000).
Experimental versus Digital Northern Analysis of Gene Expression
Large-scale single-pass sequencing projects have generated a wealth of data concerning the expression of genes in particular organisms. Databases composed of such expressed sequence tags (dbESTs) have recently been interrogated as a means of estimating relative gene expression levels (Mekhedov et al., 2000; White et al., 2000). These analyses have been termed digital or electronic northerns, and it has been suggested that EST abundance represents gene expression patterns that may be used to determine the existence of metabolic regulons (White et al., 2000).
Analysis of the plant dbESTs (Mekhedov et al., 2000) indicated that the electronic data compiled from all plant sources showed a remarkable correlation with that derived experimentally in this study. Table I presents the data from each method relative to the abundance of KR mRNA. Comparison of mRNA abundance levels for BC, KR, ENR, and TE revealed that for these genes there was no more than a 2-fold difference. In the electronic data, the relative levels of DES and ACP were approximately 2-fold lower, whereas the level of type I ACCase was 6-fold higher than when experimentally determined. This was surprising because most plant ESTs are derived largely from vegetative tissue that may under-represent the abundance of lipid synthetic mRNAs. To address this problem, White et al. (2000) recently reported a new set of Arabidopsis ESTs derived from developing seed tissue. The correlation previously seen between experimental and electronic northerns in the relative abundance levels of lipid synthetic mRNAs was not apparent in this data set. ESTs for DH and ENR were not detected at all in the seed ESTs, whereas the abundance of these mRNAs relative to KR was found experimentally to be approximately 0.5 and 1.0, respectively.
A significance test has been developed to account for the random fluctuations and sampling size on the reliability of EST data (Audic and Claverie, 1997). Using this statistical method, the 16 KR ESTs sequenced from seed tissue (White et al., 2000) cannot be distinguished from between 5 and 34 ESTs at the 99% confidence limit. Because no mRNAs for ENR were sequenced in the EST data set, this indicates that there is a significant difference between the steady-state levels of KR and ENR mRNAs in the seed EST library.
Relative Levels of FAS Component mRNAs in Embryo and Leaf Tissue
To determine the relative abundance of FAS transcripts in vegetative and oil storing tissues, we carried out quantitative northern analysis on total RNA from expanding leaves and developing embryos. RNA was extracted from the fourth true leaf of B. napus plants, 4 d after emergence. At this time the level of KR activity is almost at its maximum level (O'Hara et al., 2001). By comparison with the kinetics of mRNA accumulation and enzyme activity in embryos, we predicted that KR (and other FAS component) mRNAs would be at their maximum abundance.
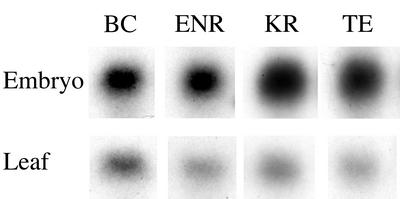
Approximately 15 μg of total RNA from each tissue was used for the analysis; the blots clearly demonstrate large differences in relative abundance of each mRNA in embryo over leaf material (Fig. 4). After adjusting for RNA loading, the numbers of FAS transcripts in leaf were calculated and compared with the maximum number of transcripts measured in embryos (Table II). The abundance of the catalytic components in embryos ranged from 3- to 15-fold greater than in leaves. Although it is not possible to make direct comparisons between different tissues, the maximum rate of fatty acid synthesis in a single leaf is 14.5 μg d−1 (O'Hara et al., 2001), whereas that of a single seed is 60 μg d−1 (determined from this embryo series; results not shown). This suggests a much greater pathway flux in the storage tissue.
Figure 4.
Relative mRNA abundance in leaf and embryo. The concentration of BC, ENR, KR, TE, and ACP mRNAs was determined by comparing the intensity of hybridization with that of gene-specific RNA concentration standards. The gel contained approximately 15 μg of total RNA from each tissue; after normalization of the signal by reprobing with an 18s rRNA sequence, the embryo to leaf signal was calculated to be: 3.4:1 (BC), 8.1:1 (ENR), 15.4:1 (KR), and 6.2:1 (TE).
Table II.
FAS component transcript abundance and ratio in embryo and leaf RNA
| FAS Component | Maximum No. of Transcripts (×106) per μg Total RNA
|
Embryo to Leaf Ratio | |
|---|---|---|---|
| Embryo | Leaf | ||
| KR | 59.6 | 3.9 | 15.4 |
| BC | 47.4 | 14 | 3.4 |
| ENR | 89.0 | 11 | 8.1 |
| TE | 29.0 | 4.7 | 6.2 |
The maximum number of transcripts detected in embryo RNA and young, expanding leaf RNA samples were used to calculate the abundance ratio in these two tissues.
The steady-state mRNA ratio of components was maintained throughout embryogenesis. However, unlike the genes encoding heteromeric ACCase (Ke et al., 2000), the ratio of FAS components in leaf RNA was different than that observed in embryo. The ratio between the mRNAs in leaf was KR:BC:ENR:TE = 1.0:3.6:2.8:1.2, representing a 6-fold increase in the ratio of BC, and 2- and 3-fold increases in the levels of ENR and TE relative to KR. The reasons for the different relative abundances of BC and FAS components in the leaf RNA is not immediately clear, but was determined from the mass of each component (Fig. 1; Table II). Analysis of heteromeric ACCase component ratios was done by a single linear regression analysis of mRNA abundance from different tissues (Ke et al., 2000), and, therefore, a single value for the ratios of the four components was determined from the slope of the regression line and applied to all tissues. Combining data from different tissues in this way may mask any tissue-specific differences in mRNA levels.
RT-PCR Analysis of Leaf KR mRNA Levels
In young, expanding leaves of B. napus the measured steady-state levels of FAS mRNAs was low. We, therefore, sought an alternative estimate of transcript abundance for one FAS component by RT-PCR.
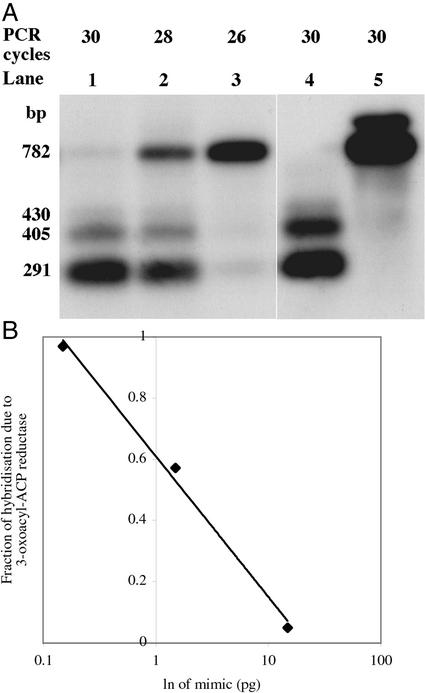
A competitive mimic of KR (KRC) was created by removal of an NdeI restriction site from clone AJ243083 and was used to spike total leaf RNA before RT reactions. Preliminary experiments were carried out to establish the number of PCR cycles that resulted in a linear increase in detectable products when adding different amounts of mimic (0.15, 1.5, or 15 pg) to a fixed quantity of total RNA. After treatment with NdeI and HpaI, which resulted in digestion of the four KR isoforms but not the KRC mimic, the RT-PCR products were separated by agarose gel electrophoresis and hybridized to a probe from the 5′ region of KR (Fig. 5A). The KRC band was 782 bp, whereas those of the KR mRNAs were 291, 405, and 430 bp. A plot of the signal detected from KR isoforms, expressed as a fraction of the total signal in the lane, plotted against the ln of the amount of mimic present, gave a linear relationship (Fig. 5B; y = −0.1998 ln[x] = 0.611; r2 = 0.994). From the equation of the regression line it was possible to determine the amount of KR mRNAs in the original sample at y = 0.5 to be 1.74 pg μg−1 leaf total RNA. Using a transcript length of 1,200 nucleotides for KR mRNA this converts to 4.4 fmol mg−1 total leaf RNA, which agrees very well with the estimate from quantitative northern analysis of 6.4 fmol mg−1 total RNA.
Figure 5.
Quantitative RT-PCR of KR mRNA in young leaf tissue. A, RT-PCR products for isoforms of KR (AJ243083–6) and a competitive mimic (KRC) were digested with NdeI and HpaI, transferred to a Zeta filter (Bio-Rad), and hybridized to a 5′ fragment of AJ243083 under conditions to ensure hybridization to all four isoforms. KRC remained undigested at 782 bp. The 5′ digestion products of AJ243084–6 ran at 405 to 430 bp and of AJ243083 at 291 bp. Lanes 1 through 4 contained PCR products from 1.0 μg of total RNA and 0.15 pg of KRC (lane 1), 1.5 pg of KRC (lane 2), 15 pg of KRC (lane 3), and no KRC control (lane 4). Lane 5 contained 15 pg of KRC only control. B, After densitometry, the integrated density of KR isoforms as a fraction of all hybridizing bands was plotted against the ln of the amount of mimic added. Linear regression of the data gave an r2 value of 0.994.
DISCUSSION
Investigations to date have suggested that major influences on the tissue and temporal modulations of ACCase and FAS component activities are the consequence of pretranslational processes. It was demonstrated recently that mRNAs encoding the four subunits of heteromeric ACCase accumulate at a constant molar ratio throughout silique development in Arabidopsis. The ratios were found to be CAC1:CAC2:CAC3:(accD-A and accD-B) = 0.14:1.0:0.17:0.06 (Ke et al., 2000). There is presently no report of the stoichiometric relationships of ACCase subunits in the mature enzyme, although it is unlikely that the ratios closely follow those of mRNA accumulation. Differences in the absolute numbers of transcripts probably reflect post-transcriptional events, e.g. mRNA stability, translation, or protein stability rather than the quaternary structure of the ACCase enzyme.
All of the FAS components tested in this study exhibited similar profiles of mRNA accumulation throughout embryogenesis. Quantitative analyses indicate that ACP mRNA shows the highest steady-state levels of all those tested. It accumulates to levels about 7-fold higher than ENR and KR and approximately twice that of DES. Given the possibility that not all ACP sequences would hybridize to the probe used, these figures should be regarded as an underestimate for ACP steady-state transcript levels. The levels of accumulation of FAS components (KR, DH, and ENR) were similar to each other, but somewhat lower than that of DES that acts upon mature acyl-ACP substrates.
Analysis of the temporal expression of G3PAT mRNA throughout embryogenesis showed a different profile to that of FAS components. There was a basal expression level throughout embryo development, similar to that found in young expanding leaves. However, there was an increase in accumulation of G3PAT mRNA between 23 and 29 DAF, before reduction to base levels by 32 DAF (results not shown). The maximum level of embryo G3PAT mRNA accumulation was only twice that seen in young expanding leaves. Although there is no known role for plastidial G3PAT in triglyceride biosynthesis, the gene responds to the same embryo cues as FAS component genes with the accumulation of higher steady-state levels of mRNA during the lipid deposition phase. The mass of G3PAT mRNA was less than the lowest in vitro generated standard used (equivalent to <5 pg 15 μg−1 total RNA) and was, therefore, 1 to 2 orders of magnitude less abundant than FAS component mRNAs.
One of the proposed uses of dbESTs is as a means of quantitative estimation of gene expression levels to aid the understanding of metabolism (Ohlrogge and Benning, 2000; White et al., 2000). Such an analysis has been carried out for lipid biosynthetic genes using the data for all plant dbESTs available at that time (Mekhedov et al., 2000). Comparison of the digital northern data with experimental quantitative steady-state mRNA analysis (Table I; Ke et al., 2000) highlights many similarities and also some inconsistencies in the electronic data. There is close agreement between digital and experimental quantification for the BC component of heteromeric ACCase (CAC2), MCAT, KR, DH, ENR, and TE. The data for DES and ACP show approximately 2-fold variation between these two methods.
Digital northern analysis showed less similarity to experimental results for the sequences of the nuclear-encoded subunits of heteromeric ACCase (CAC1, CAC2, and CAC3). These mRNAs were present in the plant EST database in a ratio of 1.6:1.0:0.6 (Mekhedov et al., 2000), whereas quantitative analysis of transcript abundance in Arabidopsis silique RNA revealed a ratio of 0.14:1.0:0.17 for these three genes (Ke et al., 2000). It is unlikely that this discrepancy arose because of differences in tissue or temporal expression because the three genes have been reported to be coordinately expressed at constant molar ratios throughout silique development and also in young expanding leaves, flower buds, and flowers (Ke et al., 2000). The differences may, therefore, arise because of artifacts in the construction/screening of EST libraries or in the quantification of RNA from northern blots.
One concern for biologists studying a tissue-specific phenomenon, such as triglyceride synthesis, is that the sequences of interest may be absent or under-represented in EST libraries synthesized largely from vegetative tissues. To remove this possibility, White et al. (2000) generated a database of approximately 10, 500 new Arabidopsis ESTs from a developing seed library; approximately 40% of the sequences had no match in dbEST and may, therefore, be seed-specific sequences. This shows the general utility of a tissue- and temporal-specific EST program, and the sequences will be a valuable resource for understanding seed metabolism. However, an unexpected imbalance in the number of ESTs that encode components of fatty acid and lipid biosynthesis is apparent in this data set (Table I; White et al., 2000). No sequences for DH or ENR were present, and those for DES and BC are under-represented when compared with all plant ESTs and our quantitative RNA-blot data. The reason for such disparate results may be due to the numbers of sequences involved in the two EST data sets. Such discrepancies mean that considerable care will be required when using database searches to make inferences about the presence or absence of metabolic routes and their regulation.
The Arabidopsis seed ESTs set has been used to make a microarray for the broad analysis of gene expression levels in developing seeds and to analyze the seed to leaf and seed to root expression ratios of many genes (Girke et al., 2000). In this analysis, 20% of lipid biosynthetic genes were expressed at ratios of ≥2-fold higher in seeds than in leaves or roots. However, inspection of the primary data highlights some major differences between the expression ratios for specific genes when compared with quantitative northern analyses. Microarray analysis of B. napus transcripts gave a seed to leaf ratio of: BC, −2; ENR, 1; KR, 1 to 2; and ACP, −2 to 2 (http://www.bpp.msu.edu/Seed/SeedArray.htm). The quantitative northern analyses presented herein show between 3- and 15-fold higher expression of FAS components in embryos over leaf tissue (Table II). The reasons for such large differences in the values of the relative abundance of these specific mRNAs from the two methods is not clear, although it may be, in part, due to the tissue source. In our experiments, embryos dissected from seeds were used, whereas the microarray analysis used seeds; the use of whole seeds may lead to an underestimate of the abundance of transcripts encoding proteins involved in storage triglyceride synthesis in the embryo.
Using quantitative mRNA analysis, we have experimentally demonstrated that several mRNAs involved in fatty acid and lipid metabolism accumulate at constant molar ratios throughout embryogenesis. Furthermore, the use of in vitro generated mRNA standards has allowed comparison of absolute levels of different transcripts.
There is good agreement in the relative transcript abundances between of the experimental and digital data in large EST collections (Mekhedov et al., 2000) but less correlation with a smaller seed-specific EST set (White et al., 2000).
The percentage representation of two ACCase components in mRNA populations has been determined independently with similar outcomes. In Arabidopsis, the maximum level of BC mRNA was calculated to be 1 mol % (Ke et al., 2000), and in B. napus, the value was found to be 0.35 mol % (this study). Similarly, two estimates of the abundance of BCCP mRNA have been reported for Arabidopsis: 0.14 mol % in siliques (Ke et al., 2000) and 0.1% to 0.5% of mRNA in developing leaves and siliques (Thelen et al., 2001). These estimates were all made using the assumption that mRNA was present as 1% of the total RNA. However, this may be an underestimate of the level of mRNA in developing embryos. In our hands, embryo RNA eluted from either oligo(dT) columns or polyATRACT mRNA isolations (Promega, Madison, WI) in zero salt represented approximately 3% of total RNA loaded (results not shown). These preparations were not checked for contamination with poly(A−) RNA, but this data may indicate that estimations of transcript representations as a percentage of all mRNAs are up to 3-fold too high.
Analysis of all plant dbEST (Mekhedov et al., 2000) identified 17 BC transcripts from a database of approximately 160,000 ESTs, an abundance of 0.01%. This represents 35-fold fewer BC transcripts registered in the electronic databases than determined experimentally in this study. Discrepancies have also been noted previously in the transcript abundance of two Arabidopsis isoforms of another subunit of the heteromeric ACCase, BCCP (AtBCCP; [Thelen et al., 2001]). The Arabidopsis EST database contained two AtBCCP1 and 11 AtBCCP2 transcripts. Analysis of transcript expression by northern blots estimated the AtBCCP1 mRNA to represent 0.1% to 0.5% of mRNA in developing leaves and that AtBCCP2 levels were approximately 4-fold lower than AtBCCP1 (Thelen et al., 2001). These data support the notion that EST libraries may be under-represented in certain sequences, possibly because of differential efficiency in the reverse transcriptase step of cDNA synthesis.
MATERIALS AND METHODS
Growth and Harvesting of Plant Material
Brassica napus (cv Westar) plants were grown in M3 Levington compost in individual 20-cm pots with a 16-h light period (20°C) and an 8-h dark period (15°C) at 60% humidity. Illumination, measured with a quantum radiometer/photometer (Macam, Livingston, Scotland), was at a photon flux density of 250 μE m−2 s−1. Flowers were tagged when fully open, and embryos were collected from 14 to 48 DAF; mature seeds were harvested when fully dry (at more than 50 DAF). All plant material was frozen in liquid nitrogen before storing at −80°C until required.
cDNA Sequences
The following cDNAs were used: homomeric ACCase (X77382); BC (AY034410); stearoyl-ACP desaturase (X63364); TE (X73849); ACP (X13128); ENR (S60064); KR (AJ243085); DH (AF382146); MCAT (AJ007046); and napin (J02586).
RNA Extraction
Total RNA were extracted from 4-d-old true leaves or 50 to 100 staged embryos, depending on developmental stage, using Trizol reagent essentially by the manufacturers protocol (Invitrogen, Carlsbad, CA). Poly(A+) mRNA was prepared from total RNA by separation on freshly prepared oligo(dT) columns (Collaborative Biomedical Products, Bedford, MA). Antisense RNA was synthesized from linearized plasmid preparations of cDNA clones of KR, BC, TE, ACP, and ENR using the Riboprobe in vitro transcription system (Promega). RNA concentrations were determined spectrophotometrically using undiluted samples. The A260 readings were between 0.1 and 1.0.
Filter Array Analysis
Alkaline denatured samples (50 and 250 ng) of each cDNA (in pBluescript SK+) were applied to Zeta-Probe GT nylon membranes using a dot-blot apparatus (Bio-Rad, Hercules, CA). Labeled first-strand cDNA was synthesized as follows: 7.0 μg of oligo(dT)15 (Boehringer Mannheim/Roche, Basel) was annealed to 0.5 to 1.0 μg of poly(A+) mRNA by heating the reaction to 70°C for 15 min and then chilling on ice.
The reaction mixture was completed to a total volume of 50 μL by adding (final concentration) 1× Superscript II buffer, 0.1 m dithiothreitol, 40 units of RNAsin (Promega); 0.5 mm each of dATP, dGTP, and dTTP and 50 μm dCTP (Amersham-Pharmacia Biotech, Uppsala); 50 μCi of [α-32P]dCTP (NEN Life Science Products, Boston); 600 units of Superscript II RNase H-reverse transcriptase (Invitrogen); and incubated at 42°C for 60 min. At the end of first-strand synthesis, the reaction was stopped by heating at 70°C for 20 min and RNA removed by incubating with 5 units of RNaseH at 37°C for 15 min. For second-strand synthesis, 16 μg of random hexanucleotide primers (Boehringer Mannheim/Roche) was added before heat denaturation. The labeling reaction was completed by the addition of 1× Klenow buffer, dNTPs (equimolar mixture) to a final concentration of 0.2 mm, 10 units of Klenow (Boehringer Mannheim/Roche), 50 μCi of [α-32P]dCTP, and incubated at 37°C for 3h or overnight. Unincorporated activity was removed by chromatography on a Biogel P-6 microspin column (Bio-Rad). Incorporation was typically of the order 104 to 105 d.p.m. after first-strand synthesis and 107 to 108 d.p.m. after random priming. RT-PCR and alkaline gel electrophoresis was used to check the effectiveness of first-strand synthesis.
Filters were prehybridized with linear denatured empty pBluescript SK+ vector (>200 μg per filter) and 5 μg of denatured poly(dA)-(dT) to reduce nonspecific binding. The probe was hybridized with the filters in fresh buffer after denaturation in a boiling bath at 100°C for 10 min and brief chilling on ice. All hybridizations were made at 65°C, and post-hybridization washes were made at high stringency after the Zeta-Probe protocol.
After removal of the first probe, the filters were rehybridized with a β-lactamase DNA sequence, labeled using the Rediprime DNA labeling system (Amersham-Pharmacia Biotech), to determine the equivalence of sample loading and DNA transfer to the filter. In each plasmid, β-lactamase was present at the same copy number as the insert.
Northern Analysis
Gel preparation and northern blotting was carried out by standard procedures (Sambrook et al., 1989). Total RNA (approximately 15 μg) from each time point in the embryo series and for developing leaves was electrophoresed through a denaturing gel before blotting to a Hybond N (Amersham-Pharmacia Biotech) nylon membrane. For quantitative analysis, dilutions of antisense RNA (0.005–1 ng) were loaded into a second series of wells before electrophoresis was completed. Probes were prepared from 25 ng of purified insert from each cDNA and labeled using the Rediprime DNA labeling system (Amersham-Pharmacia Biotech). Unincorporated activity was removed by chromatography on a Biogel P-6 microspin column.
Hybridization and Washing Conditions
Hybridization was performed in (5× SSPE, 5× Denhardt, 0.5% [w/v] SDS, 50% [v/v] formamide, and 20 μg mL−1 herring sperm DNA) at 42°C overnight. Membranes were then washed (twice in 2× SSC, 0.1% [w/v] SDS at 25°C for 10 min; once in 1× SSC, 0.1% [w/v] SDS at 50°C for 15 min; and twice in 0.1× SSC and 0.1% [w/v] SDS at 50°C for 10 min).
The mole fraction G plus C composition, monovalent cation concentration, the length of the probe, and the sequence similarity of the probe to the target determine whether a hybridizing signal is detected. Using the relationship determined by Meinkoth and Wahl (Meinkoth and Wahl, 1984) and the mole fraction G plus C residues of 47.6% for the coding region of the B. napus genome (http://www.kazusa.or.jp), we determined a melting temperature of 70.9°C for a 1-kb probe in 0.0165 Na+. Using the rule that a 1% mismatch results in a drop in melting temperature of 1°C (Meinkoth and Wahl, 1984), the washes used would remove hybrids with less than approximately 79% identity.
Competitive RT-PCR
KR transcript abundance in leaf total mRNA was determined by RT-PCR. A KR mimic (KRc) was produced from the AJ243083 sequence by filling in the NdeI site cohesive ends after digestion with this enzyme and blunt-end ligation.
Approximately 30 μg of total leaf RNA was incubated with 2 units of RQ1 DNase (Promega) at 37°C for 20 min. The LiCl-precipitated pellet was washed twice in 70% (v/v) ethanol, and 25 μg was recovered in 40 μL of sterile distilled water. The RT reaction was primed using a consensus sequence from the 3′-untranslated region of the KR isoforms (5′-AACAGAAATCCGACCAAGTGCCAGA-3′). Primer and total RNA were first prepared as a cocktail and aliquots, containing 1.0 μg of total RNA, and 20 pmol of primer was then spiked with 0.15, 1.5, or 15 pg of sense KRc mRNA. RT reactions were as previously described.
PCR components were prepared as a cocktail to which 2.0 μL of each RT reaction was added. The primers conformed to consensus sequences in the four isoforms of KR. Forward primer, 5′-GTGAGATCCGTCAGGTCCGTCAATGG-3′; reverse primer, 5′-CTACCAAGCCAGCCACATCTTCAGG-3′. The PCR parameters were one cycle at 94°C for 5 min and 24 to 30 cycles at 94°C for 40 s, 62°C for 1 min, and 72°C for 1 min and 30 s. Only reactions that showed a linear response with increased cycles were used in subsequent analyses.
Ethanol-precipitated products were digested with NdeI (site in AJ243083) and HpaI (sites in AJ243084–6, but absent in AJ243083 and KRC) before separation on a 2% (w/v) agarose gel. DNA fragments were alkaline transferred onto a nylon membrane and UV cross-linked. The filter was hybridized with an EcoRI/NdeI fragment from the 5′ end of the AJ243083, labeled as previously described. Prehybridization and hybridization was done as described previously, except that no formamide was included in the buffer, and hybridization was performed at 65°C. The filter was washed (twice with 2× SSC, 0.1% [w/v] SDS at 25°C for 10 min; and once with 1× SSC, 0.1% [w/v] SDS at 48°C for 15 min). At this stringency, all KR isoforms hybridized to the probe.
Data Analysis
Filter arrays and northern blots were analyzed using a Molecular Imager System and quantification performed with the Molecular Analyst (Bio-Rad) software. Results for northern blots were normalized after reprobing with a heterologous 18S rDNA sequence from pea (Pisum sativum). Transcripts were quantified by reference to a regression line fitted to the corresponding antisense dilution series present on the same blot.
Footnotes
This work was supported by the Biotechnology and Biological Science Research Council under the Resource Allocation and Stress in Plants initiative (grant no. RSP 07674).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010956.
LITERATURE CITED
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Baerson SR, Vanderheiden MG, Lamppa GK. Identification of domains in an Arabidopsis acyl carrier protein gene promoter required for maximal organ-specific expression. Plant Mol Biol. 1994;26:1947–1959. doi: 10.1007/BF00019505. [DOI] [PubMed] [Google Scholar]
- Caughey I, Kekwick RGO. The characteristics of some components of the fatty-acid synthetase system in the plastids from the mesocarp of avocado (Persea americana) fruit. Eur J Biochem. 1982;123:553–561. doi: 10.1111/j.1432-1033.1982.tb06568.x. [DOI] [PubMed] [Google Scholar]
- de Boer GJ, Kater MM, Fawcett T, Slabas AR, Nijkamp HJJ, Stuitje AR. The NADH-specific enoyl acyl carrier protein reductase: characterization of a housekeeping gene involved in storage lipid synthesis in seeds of Arabidopsis and other plant species. Plant Physiol Biochem. 1998;36:473–486. [Google Scholar]
- de Boer GJ, Testerink C, Pielage G, Nijkamp HJJ, Stuitje AR. Sequences surrounding the transcription initiation site of the Arabidopsis enoyl-acyl carrier protein reductase gene control seed expression in transgenic tobacco. Plant Mol Biol. 1999;39:1197–1207. doi: 10.1023/a:1006129924683. [DOI] [PubMed] [Google Scholar]
- de Silva J, Loader NM, Jarman C, Windust JHC, Hughes SG, Safford R. The isolation and sequence-analysis of 2 seed-expressed acyl carrier protein genes from Brassica napus. Plant Mol Biol. 1990;14:537–548. doi: 10.1007/BF00027499. [DOI] [PubMed] [Google Scholar]
- Elborough KM, Swinhoe R, Winz R, Kroon JTM, Farnsworth L, Fawcett T, Martinezrivas JM, Slabas AR. Isolation of cDNAs from Brassica napus encoding the biotin-binding and transcarboxylase domains of acetyl-CoA carboxylase: assignment of the domain-structure in a full-length Arabidopsis thaliana genomic clone. Biochem J. 1994;301:599–605. doi: 10.1042/bj3010599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T, Simon WJ, Swinhoe R, Shanklin J, Nishida I, Christie WW, Slabas AR. Expression of messenger-RNA and steady-state levels of protein isoforms of enoyl-ACP reductase from Brassica napus. Plant Mol Biol. 1994;26:155–163. doi: 10.1007/BF00039528. [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlousek-Radojcic A, Postbeittenmiller D, Ohlrogge JB. Expression of constitutive and tissue-specific acyl carrier protein isoforms in Arabidopsis. Plant Physiol. 1992;98:206–214. doi: 10.1104/pp.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoj PB, Mikkelsen JD. Partial separation of individual enzyme-activities of an ACP-dependent fatty-acid synthetase from barley chloroplasts. Carlsberg Res Commun. 1982;47:119–141. [Google Scholar]
- Ke JS, Wen TN, Nikolau BJ, Wurtele ES. Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme a carboxylase. Plant Physiol. 2000;122:1057–1071. doi: 10.1104/pp.122.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J, Wahl G. Hybridization of nucleic-acids immobilized on solid supports. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Mekhedov S, de Ilarduya OM, Ohlrogge J. Toward a functional catalog of the plant genome: a survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–401. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Cummins I. Biosynthesis of seed storage products during embryogenesis in rapeseed, Brassica napus. J Plant Physiol. 1989;135:63–69. [Google Scholar]
- O'Hara P, Slabas AR, Fawcett T. Fatty acid synthesis in developing leaves of Brassica napus in relation to leaf growth and changes is activity of 3-oxoacyl-ACP reductase. FEBS Lett. 2001;488:18–22. doi: 10.1016/s0014-5793(00)02406-6. [DOI] [PubMed] [Google Scholar]
- Ohlrogge J, Benning C. Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol. 2000;3:224–228. [PubMed] [Google Scholar]
- Safford R, Windust JHC, Lucas C, de Silva J, James CM, Hellyer A, Smith CG, Slabas AR, Hughes SG. Plastid-localized seed acyl-carrier protein of Brassica napus is encoded by a distinct, nuclear multigene family. Eur J Biochem. 1988;174:287–295. doi: 10.1111/j.1432-1033.1988.tb14096.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki Y, Hakamada K, Suama Y, Nagano Y, Furusawa I, Matsuno R. Chloroplast-encoded protein as a subunit of acetyl-CoA carboxylase in pea plant. J Biol Chem. 1993;268:25118–25123. [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y. The compartmentation of acetyl-coenzyme-a carboxylase in plants. Plant Physiol. 1995;108:445–449. doi: 10.1104/pp.108.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage LJ, Ohlrogge JB. Phosphorylation of pea chloroplast acetyl-CoA carboxylase. Plant J. 1999;18:521–527. doi: 10.1046/j.1365-313x.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- Shimakata T, Stumpf PK. The procaryotic nature of the fatty-acid synthetase of developing Carthamus tinctorius L (safflower) seeds. Arch Biochem Biophys. 1982;217:144–154. doi: 10.1016/0003-9861(82)90488-x. [DOI] [PubMed] [Google Scholar]
- Slocombe SP, Cummins I, Jarvis RP, Murphy DJ. Nucleotide sequence and temporal regulation of a seed-specific Brassica napus cDNA encoding a stearoyl-acyl carrier protein (ACP) desaturase. Plant Mol Biol. 1992;20:151–155. doi: 10.1007/BF00029157. [DOI] [PubMed] [Google Scholar]
- Slocombe SP, Piffanelli P, Fairbairn D, Bowra S, Hatzopoulos P, Tsiantis M, Murphy DJ. Temporal and tissue-specific regulation of a Brassica napus stearoyl-acyl carrier protein desaturase gene. Plant Physiol. 1994;104:1167–1176. doi: 10.1104/pp.104.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Mekhedov S, Ohlrogge JB. Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol. 2001;125:2016–2028. doi: 10.1104/pp.125.4.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Todd T, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C. A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]