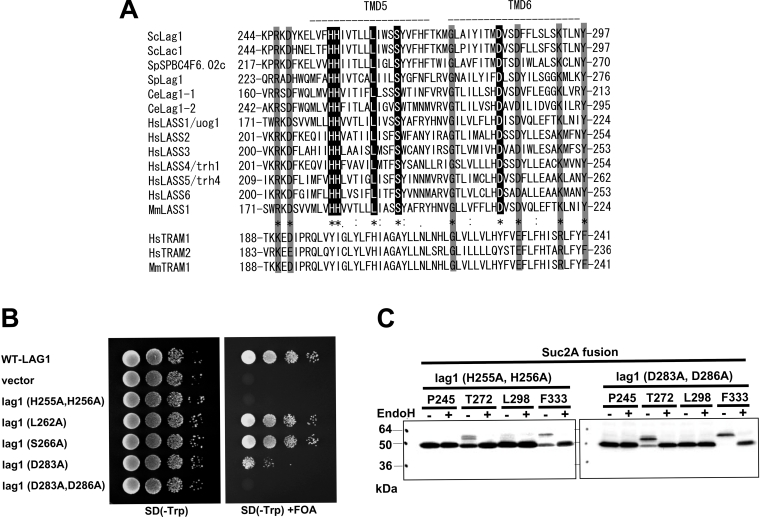
Figure 5. Alignment of the Lag motif.
(A) Alignment of amino acid sequences of the Lag motif of Lag1 homologues and TRAM. The CLUSTALW algorithm was used to create the sequence alignment. The ‘*’ indicates positions that have a single, fully conserved residue among Lag1 homologues. The ‘:’ and ‘.’ indicate that the stronger- and weaker-score groups are fully conserved among Lag1 homologues respectively. Sc, S. cerevisiae; Sp, S. pombe; Ce, Caenorhabditis elegans; Hs, H. sapiens; Mm, Mus musculus. Note that some amino acid residues showed similarity in both Lag1 homologues and TRAM (shaded box) homologues, but others are conserved only among Lag1 homologues (black box). (B) Mutation analysis of the conserved amino acids among Lag1 homologues but not in TRAM. lag1Δ lac1Δ double deletion mutant cells transformed with pRS416-LAG1 (LAG1 on uracil-based plasmid) (RH6602) were transformed with pRS414-lag1(H255A/H256A), pRS414-lag1(L262A), pRS414-lag1(S266A), pRS414-lag1(D283A) and pRS414-lag1(D283A/D286A) (plasmids 1825–1829). The transformants were tested for complementation as in Figure 2(B). (C) H255A/H256A and D283A/D286A mutations were introduced into the LAG1-SUC2A chimaeric constructs used in Figure 2(B), resulting in plasmids 1853–1860. Protein expression and detection were performed as described in Figure 2(A).