Abstract
Peters Plus syndrome is an autosomal recessive disorder characterized by anterior eye-chamber abnormalities, disproportionate short stature, and developmental delay. After detection of a microdeletion by array-based comparative genomic hybridization, we identified biallelic truncating mutations in the β1,3-galactosyltransferase–like gene (B3GALTL) in all 20 tested patients, showing that Peters Plus is a monogenic, primarily single-mutation syndrome. This finding is expected to put Peters Plus syndrome on the growing list of congenital malformation syndromes caused by glycosylation defects.
Peters Plus syndrome (MIM 261540) is an autosomal recessive disorder characterized by a variety of anterior eye-chamber defects, of which the Peters anomaly occurs most frequently.1 Other major symptoms are a disproportionate short stature, developmental delay, characteristic craniofacial features, and cleft lip and/or palate.1
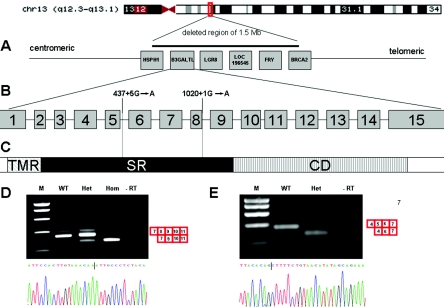
To detect potential microrearrangements affecting the disease locus, we performed genomewide 1-Mb resolution array-based comparative genomic hybridization2 on genomic DNA of two brothers and four isolated patients who all received the clinical diagnosis of Peters Plus syndrome. In both brothers, two adjacent BAC clones (RP11-95N14 and RP11-37E23) were found to be present in a single copy, representing an ∼1.5-Mb interstitial deletion on chromosome 13 (q12.3q13.1). MLPA (multiplex ligation-dependent probe amplification) analysis was used to confirm the deletion and to better define its extent. The deletion was confirmed in both brothers and their mother and spans six genes (HSPH1, B3GALTL, LGR8, LOC196545, FRY, and the first 13 exons of the BRCA2 gene). Two of these, LGR8 and BRCA2, are associated with human disease. Mutations in LGR8 cause testicular maldescent3; since both brothers had cryptorchidism, this may be related to their LGR8 haploinsufficiency. BRCA2 mutations are associated with hereditary breast and ovarian cancer, and large genomic rearrangements are known to contribute to ∼2% of the BRCA2 mutation spectrum.4,5 The brothers’ family history was positive for breast cancer in at least two deceased female relatives, in whom we established the presence of the deletion by interphase FISH on tumor material. Thus, this deletion constitutes a novel large BRCA2 rearrangement associated with familial breast cancer.
Since none of the six genes was an obvious candidate gene for Peters Plus syndrome, we sequenced the genes' exons and flanking sequences in one of the affected brothers. A point mutation (c.1020+1G→A) was detected in the β1,3-galactosyltransferase–like gene (HUGO Gene Nomenclature Committee symbol B3GALTL) within the donor splice site of exon 8. The same mutation was also present in the other brother and as a single copy in the father. We subsequently performed targeted sequencing analysis for the presence of the c.1020+1G→A mutation in an additional 18 patients with Peters Plus from 15 families. Fourteen patients were Dutch whites, and the other patients were Turkish, British, Arab, or Indian. All had the salient features of Peters Plus syndrome (table 1). We detected a homozygous c.1020+1G→A mutation in 16 of the 18 patients. In the remaining two patients (Dutch siblings), only a single c.1020+1G→A mutation was present (on the maternal allele). On sequencing the remainder of the gene, we detected a point mutation in intron 5 of B3GALTL (c.437+5G→A) on the paternal allele. Of the 11 available parent sets, all were heterozygous for the mutation detected in their affected offspring. We then excluded the presence of the c.1020+1G→A and c.437+5G→A mutations in 455 chromosomes of healthy Dutch individuals, by melting-curve analysis with specifically designed primer sequences (LightScanner HR96 [Idaho Technology]). Also, we investigated whether c.1020+1G→A could be a founder mutation, by analyzing known intragenic B3GALTL SNPs in 18 of the homozygous patients. Seven patients (Italian, Turkish, English, and four Dutch) showed heterozygosity for at least one of the three informative SNPs (rs9315120, rs877103, and rs877104 [dbSNP]), which indicates that it is most likely a recurrent mutation, although some of the Dutch patients may have a common ancestor. The mutation is at the site of a potentially methylated CpG dinucleotide, which could explain its recurrence.6
Table 1. .
Clinical Characteristics of Individuals with Peters Plus Syndrome and Mutations of B3GALTL[Note]
| Individual | Sex | Peters Anomaly | Anterior Eye-Chamber Anomaly | Disproportionate Short Staturea | Cleft Lip and/or Palate |
Developmental Delay | Heart Anomaly | Renal Anomaly | Ethnic Origin | Mutation |
| 1100.1 | F | − | + | + | − | + | − | + | Dutch | Homozygous 1020+1G→A |
| 1100.2b | M | − | + | + | − | U | − | − | Dutch | Homozygous 1020+1G→A |
| 1200.1 | F | + | + | + | − | + | − | − | Dutch | Homozygous 1020+1G→A |
| 1200.2 | F | + | + | + | − | + | − | − | Dutch | Homozygous 1020+1G→A |
| 1201.5 | F | + | + | + | L | + | − | − | Dutch | 1020+1G→Amat/437+5G→Apat |
| 1201.6 | M | + | + | + | − | + | − | − | Dutch | 1020+1G→Amat/437+5G→Apat |
| 1300.1 | F | + | + | + | L/P | + | − | − | Dutch | Homozygous 1020+1G→A |
| 1400.2 | M | − | + | + | L/P | + | − | − | Dutch | Homozygous 1020+1G→A |
| 1500.1 | M | + | + | + | BL/P | + | − | − | Turkish | Homozygous 1020+1G→A |
| 1600.1 | M | + | + | + | P | + | + | − | Dutch | 1020+1G→Apat/delmat |
| 1600.2 | M | U | + | + | L/P | + | + | − | Dutch | 1020+1G→Apat/delmat |
| 1700.1 | F | − | + | + | BL/P | + | + | − | Dutch | Homozygous 1020+1G→A |
| 1800.1 | M | + | + | + | − | + | − | − | Dutch | Homozygous 1020+1G→A |
| 1900.1 | F | + | + | + | − | − | − | − | Dutch | Homozygous 1020+1G→A |
| 1900.2 | M | + | + | + | − | − | − | − | Dutch | Homozygous 1020+1G→A |
| 2000.1 | F | + | + | + | L | + | + | − | Dutch | Homozygous 1020+1G→A |
| 2100.1 | M | + | + | + | − | − | − | − | Dutch | Homozygous 1020+1G→A |
| 2200.1 | M | + | + | + | BL/P | + | − | + | English | Homozygous 1020+1G→A |
| 2400.1 | F | + | + | + | − | − | + | − | Arab | Homozygous 1020+1G→A |
| 2500.1 | M | + | + | + | − | + | U | U | Indian | Homozygous 1020+1G→A |
Note.— L = cleft lip; P = cleft palate; L/P = unilateral cleft lip and palate; BL/P = bilateral cleft lip and palate; U = unknown.
<3rd Percentile.
Deceased in neonatal period.
A deleterious effect of the c.1020+1G→A mutation on transcription is certain, since it alters a donor splice site that is predicted to produce a skip of exon 8 and an out-of-frame mRNA product. We verified this by RT-PCR on patient material (fig. 1D). The c.437+5G→A mutation changes a highly conserved nucleotide and is predicted to affect splicing (Berkeley Drosophila Genome Project). To confirm this, we performed an RT-PCR on RNA isolated from lymphocytes from a patient with Peters Plus syndrome (c.1020+1G→Amat/c.437+5G→Apat). The patient's cDNA showed a skipped band, lacking exon 5, that results in an out-of-frame product. Notably, the expression of this band is much higher than that of the faint wild-type (WT) band, which is the product of the allele carrying the c.1020+1G→A mutation in exon 8 (fig. 1E). An explanation may be that the transcript lacking exon 8 is unstable. This theory is compatible with the fact that the individual who is heterozygous for the c.1020+1G→A mutation (fig. 1D [Het]), also shows a low expression of this product.
Figure 1. .
Overview of the location of the mutations in the B3GALTL gene and the results of the RT-PCR of RNA isolated from fibroblasts. A, Genes present in the 1.5-Mb deletion found in two brothers with Peters Plus syndrome. B, 15 exons of the B3GALTL gene, with the localization of the mutations. C, B3GALTL protein, which consists of a transmembrane region (TMR), a stem region (SR), and a catalytic domain (CD). Both mutations (c.1020+1G→A and c.437+5G→A) are located in the stem region. D, Result of the nested RT-PCR of exons 7–11 of the BGALTL gene, with RNA derived from myoblasts (WT), RNA from fibroblasts of a father heterozygous for the c.1020+1G→A mutation (Het), and RNA from fibroblasts of his affected son with c.1020+1G→Apat/delmat (Hom). The patient shows a smaller band compared with the WT band, which indicates a skip of exon 8. Sequence analysis of this band is shown. The vertical line indicates the end of exon 7 and the beginning of exon 9. The RT-PCR of the father shows, in addition to the WT band, a skipped product with much less intensity. E, Result of the RT-PCR encompassing exons 4–7 of the BGALTL gene, with RNA derived from lymphocytes of a control individual (WT) and a patient with a c.1020+1G→Amat/c.437+5G→Apat genotype (Het). In addition to a faint WT band, the patient shows a smaller product that lacks exon 5. The sequence analysis of this smaller band confirms the skip of exon 5.
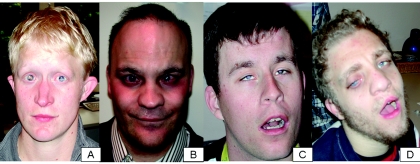
B3GALTL contains 15 exons and spans 132 kb of genomic DNA. It is transcribed in a wide range of human tissues (dbEST Web site), in the form of two transcripts (of 4.2 kb and 3.4 kb), and there is evidence of strong tissue or cell type–specific regulation.7 Transcription has been shown to terminate at three different alternative polyA-addition sites, all in exon 15.7 The B3GALTL protein spans 498 aa and contains a short N-terminal tail, a transmembrane region (aa 5–28), a so-called stem region (aa 29–260), and a C-terminal catalytic domain (aa 261–498).7 On the basis of the sequence of its catalytic domain, the protein most closely resembles proteins from the GT31 family of beta-3 glycosyltransferases (CAZy [Carbohydrate-Active enZymes Web site]). Both the c.1020+1G→A and the c.437+5G→A mutations in B3GALTL are predicted to lead to a truncated product lacking the catalytic domain, since they are located in the putative stem region of the protein (fig. 1C).7 Thus, since all patients we analyzed have homozygous severely truncating mutations, it is expected that they have, effectively, full knockout mutations and lack any significant B3GALTL activity. Given this genetic homogeneity, there is a strikingly variable cognitive phenotype. Even within the group homozygous for the c.1020+1G→A mutation, patients range from having normal secondary education to severe cognitive impairment, which suggests that other factors modulate the phenotype. The brothers with the deletion of one of their alleles (c.1020+1G→Apat/delmat) have severe cognitive impairment that is within the range of Peters Plus syndrome, and they have no structural malformations outside the Peters Plus spectrum. This indicates that hemizygosity for the genes HSPH1, LOC196545, and FRY, which have hitherto not been associated with human congenital malformations, did not produce a detectable phenotype. Figure 2 illustrates the facial phenotypes of four patients with Peters Plus syndrome.
Figure 2. .
Facial features of four patients with Peters Plus syndrome. Patients A and C are homozygous for the c.1020+1G→A mutation. Patient B has the c.1020+1G→Amat/c.437+5G→Apat genotype, and patient D has the c.1020+1G→Apat/delmat genotype. Note the Peters anomaly of the eyes, the long face, and the Cupid’s bow shape of the upper lip in all patients. Patients B and D have a repaired cleft lip and/or palate. Patient A is female; the rest are male.
B3GALTL is a putative glycosyltransferase that has not been previously associated with human disease or congenital malformations but has recently been shown to be overexpressed in thyroid oncocytic tumors.8 So far, we have not been able to verify a glycosylation defect in patients with Peters Plus syndrome; serum transferrin isoelectric-focusing studies in six of the current patients had normal results. We also studied profiles of enzymatically released N-glycans by matrix-assisted laser-desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) and high-pH anion-exchange chromatography (HPAEC) with electrochemical detection. No obvious differences in overall N-glycosylation of serum proteins were observed (results not shown). However, these results do not exclude a glycosylation defect,9 and we are initiating further (functional) studies.
There are several hundred glycosyltransferases, predicted to be active in humans, that are involved in the posttranslational modification of proteins by the addition of specific oligosaccharide side chains (glycans), to form glycoproteins. Congenital disorders of glycosylation are due to defects in the synthesis of the glycan moiety of glycoproteins or other glycoconjugates.10 Mutations in a number of glycosyltransferases have been associated with congenital malformation syndromes.10 Pending confirmation of the glycosylation defect, Peters Plus syndrome can most likely be added to this growing list. Anterior eye-chamber defects, such as Peters eye anomaly and glaucoma, are also described in Walker-Warburg syndrome and muscle-eye-brain disease,10,11 which suggests that adequate glycosylation plays a critical role in the formation of the anterior eye chamber.11,12 Interestingly, at least one Peters Plus–affected family in the present study has a documented history of glaucoma in confirmed mutation carriers. This raises the question of whether haploinsufficiency of—and possibly variations in—B3GALTL increases glaucoma susceptibility, which warrants further research. Finally, the present study emphasizes the value of genomewide array analysis in establishing the genetic basis of autosomal recessive disorders.
Acknowledgments
We thank the patients and their families for their generous cooperation, and we thank the following clinicians for referral of patients: J. van der Smagt (The Netherlands), I. C. Verma (India), L. Basel-Vanagaite (Israel), D. Bartholdi (Switzerland), and L. Wilson (United Kingdom). We also thank H. C. Hokke and A. M. Deelder (Biomolecular Mass Spectrometry Unit, Leiden), for glycosylation analysis; B. J. Poorthuis, for performing isoelectric-focusing studies; J. Knijnenburg and R. Vossen (Leiden Genome Technology Centre), for technical assistance; and A. Aartsma-Rus, for expert advice regarding the RT-PCR.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Berkeley Drosophila Genome Project, http://www.fruitfly.org/seq_tools/splice.html (for the Splice Site Prediction by Neural Network)
- Carbohydrate-Active enZymes (CAZy), http://194.214.212.50/CAZY/fam/GT31.html
- dbEST, http://www.ncbi.nlm.nih.gov/dbEST/ (for the Expressed Sequence Tags database)
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNP identification numbers rs9315120, rs877103, and rs877104)
- HUGO Gene Nomenclature Committee, http://www.gene.ucl.ac.uk/nomenclature/ (for B3GALTL)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Peters Plus syndrome)
References
- 1.Wenniger-Prick LJJM, Hennekam RCM (2002) The Peters’ plus syndrome: a review. Ann Genet 45:97–103 [DOI] [PubMed] [Google Scholar]
- 2.Knijnenburg J, Szuhai K, Giltay J, Molenaar L, Sloos W, Poot M, Tanke HJ, Rosenberg C (2005) Insights from genomic microarrays into structural chromosome rearrangements. Am J Med Genet A 132:36–40 [DOI] [PubMed] [Google Scholar]
- 3.Ferlin A, Simonato M, Bartoloni L, Rizzo G, Bettella A, Dottorini T, Dallapiccola B, Foresta C (2003) The INSL3-LGR8/GREAT ligand-receptor pair in human cryptorchidism. J Clin Endocrinol Metab 88:4273–4279 10.1210/jc.2003-030359 [DOI] [PubMed] [Google Scholar]
- 4.Tournier I, Paillerets BB, Sobol H, Stoppa-Lyonnet D, Lidereau R, Barrois M, Mazoyer S, Coulet F, Hardouin A, Chompret A, Lortholary A, Chappuis P, Bourdon V, Bonadona V, Maugard C, Gilbert B, Nogues C, Frebourg T, Tosi M (2004) Significant contribution of germline BRCA2 rearrangements in male breast cancer families. Cancer Res 64:8143–8147 10.1158/0008-5472.CAN-04-2467 [DOI] [PubMed] [Google Scholar]
- 5.Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, Mandell J, Lee MK, Ciernikova S, Foretova L, Soucek P, King MC (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295:1379–1388 10.1001/jama.295.12.1379 [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z, Zhang F (2006) Sequence context analysis of 8.2 million single nucleotide polymorphisms in the human genome. Gene 366:316–324 10.1016/j.gene.2005.08.024 [DOI] [PubMed] [Google Scholar]
- 7.Heinonen TYK, Pasternack L, Lindfors K, Breton C, Gastinel LN, Maki M, Kainulainen H (2003) A novel human glycosyltransferase: primary structure and characterization of the gene and transcripts. Biochem Biophys Res Commun 309:166–174 10.1016/S0006-291X(03)01540-7 [DOI] [PubMed] [Google Scholar]
- 8.Jacques C, Baris O, Prunier-Mirebeau D, Savagner F, Rodien P, Rohmer V, Franc B, Guyetant S, Malthiery Y, Reynier P (2005) Two-step differential expression analysis reveals a new set of genes involved in thyroid oncocytic tumors. J Clin Endocrinol Metab 90:2314–2320 10.1210/jc.2004-1337 [DOI] [PubMed] [Google Scholar]
- 9.Freeze HH (2006) Genetic defects in the human glycome. Nat Rev Genet 7:537–551 10.1038/nrg1894 [DOI] [PubMed] [Google Scholar]
- 10.Jaeken J (2003) Komrower lecture: congenital disorders of glycosylation (CDG): it’s all in it! J Inherit Metab Dis 26:99–118 10.1023/A:1024431131208 [DOI] [PubMed] [Google Scholar]
- 11.van Reeuwijk J, Janssen M, van den EC, Beltran-Valero de Bernabe D, Sabatelli P, Merlini L, Boon M, Scheffer H, Brockington M, Muntoni F, Huynen MA, Verrips A, Walsh CA, Barth PG, Brunner HG, van Bokhoven H (2005) POMT2 mutations cause α-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet 42:907–912 10.1136/jmg.2005.031963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diskin S, Kumar J, Cao Z, Schuman JS, Gilmartin T, Head SR, Panjwani N (2006) Detection of differentially expressed glycogenes in trabecular meshwork of eyes with primary open-angle glaucoma. Invest Ophthalmol Vis Sci 47:1491–1499 10.1167/iovs.05-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]