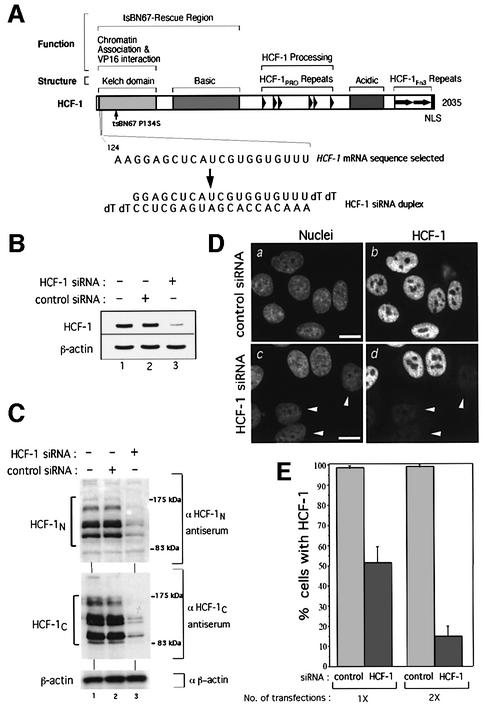
Fig. 1. Human HCF-1 is depleted efficiently and specifically by siRNA. (A) Schematic structure of HCF-1 (Wysocka et al., 2001a). Above are shown (i) structural or sequence elements and (ii) functional regions. The position of the tsBN67 P134S missense mutation is indicated. The selected HCF-1 coding region RNA sequence (nucleotides 124–144; Wilson et al., 1993) and the corresponding HCF-1 siRNA duplex are indicated below. (B) HCF-1 (top) and β-actin (bottom) mRNA levels in untreated HeLa cells (lane 1) or 3 days after treatment with either control (lane 2) or HCF-1 (lane 3) siRNA. (C) Immunoblot analysis of HCF-1N (top), HCF-1C (middle) and β-actin (bottom) proteins of untreated HeLa cells (lanes 1) or 3 days after treatment with control (lanes 2) or HCF-1 (lanes 3) siRNA. (D) Control (a and b) or HCF-1 (c and d) siRNA-treated cells were subjected to analysis after paraformaldehyde fixation by nuclear DAPI (a and c) and αHCF-1N (b and d) staining. Arrowheads point to cells exhibiting HCF-1 depletion. Scale bar: 5 µm. (E) Percentage of HCF-1-positive cells after one or two transfections with control or HCF-1 siRNA as determined by immunofluorescence. The percentage of HCF-1-positive nuclei was obtained by counting 200 cells per sample in each of three independent experiments.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.