Abstract
DNA barcodes can provide rapid species identification and aid species inventories in taxonomically unstudied groups. However, the approach may fail in recently diverged groups with complex gene histories, such as those typically found on oceanic islands. We produced a DNA-based inventory of taxonomically little known diving beetles (genus Copelatus) in the Fiji archipelago, where they are a dominant component of the aquatic invertebrate fauna. Sampling from 25 localities on five islands and analysis of sequences from one nuclear (328 bp histone 3) and three mitochondrial (492 bp rrnL, 786 bp cox1, 333 bp cob) gene regions revealed high haplotype diversity, mainly originated since the Pleistocene, and subdivided into three major phylogenetic lineages and 22 statistical parsimony networks. A traditional taxonomic study delineated 25 morphologically defined species that were largely incongruent with the DNA-based groups. Haplotype diversity and their spatial arrangement demonstrated a continuum of relatedness in Fijian Copelatus, with evidence for introgression at various hierarchical levels. The study illustrates the difficulties for formal classification in evolutionarily complex lineages, and the potentially misleading conclusions obtained from either DNA barcodes or morphological traits alone. However, the sequence profile of Fijian Copelatus provides an evolutionary framework for the group and a DNA-based reference system for the integration of ecological and other biodiversity data, independent of the Linnaean naming system.
Keywords: Copelatus, species, DNA barcoding, Fiji, taxonomy, tropical biodiversity
1. Introduction
The limited knowledge of species diversity in many areas of the globe, coupled with anthropogenic disturbance of ecosystems, has prompted calls for an improved system of inventorying biodiversity and disseminating taxonomic information (Wilson 2003; Blaxter 2004; Janzen 2004; Seberg 2004). With increasing automation of sample analysis, biological inventories can now be conducted using ‘DNA barcodes’ from segments of mitochondrial or nuclear loci (Hebert et al. 2003a; Proudlove & Wood 2003; Tautz et al. 2003; Monaghan et al. 2005). These inventories can be used to estimate genetic variation and species diversity even where no prior taxonomic analysis is available. Barcodes as currently applied are species identifiers, assuming that DNA variation within species is much lower (10× or less) than between species (Hebert et al. 2004). However, the utility of this approach remains to be tested more broadly, in particular in biologically complex situations where lineages are composed of closely related species or are affected by a complicated evolutionary history of gene trees (Moritz & Cicero 2004; Will & Rubinoff 2004).
Islands are of particular conservation and scientific interest in the global inventory of biodiversity. They often harbour unique, recently diversified faunas and floras that can present a particular challenge to taxonomic study (Mayr 1963; Gillespie & Roderick 2002; Shaw 2002; Ennos et al. 2005). The islands of the South Pacific are known to harbour large radiations of invertebrates with sometimes complex relationships to mainland taxa and other islands (Zink 1991; Clarke et al. 1996; Keast & Miller 1997; Paulian 1998; Joy & Conn 2001). Many of these groups remain poorly investigated, while pressure on their natural habitats steadily increases (Dirzo & Raven 2003).
Here, we generated a sequence-based profile of species diversity for a virtually unknown, but taxonomically complex group of organisms in the islands of Fiji, one of the larger archipelagos in the South Pacific. A recent survey of Fiji's aquatic ecosystems (M. Balke & G. Wewalka 2003, unpublished work) revealed an abundance of predaceous diving beetles (Dytiscidae) in the genus Copelatus. Based on their morphological differences, they represent numerous closely related species. To date, only five species of Copelatus have been described from Fiji, of which only C. fidschiensis (Zimmermann 1928) can be recognized with some confidence, due to ambiguous original descriptions and unavailability of the type material. We surveyed communities of Copelatus throughout the archipelago in order to assess DNA variation of the entire assemblage, followed by examination of the morphological characters traditionally used to delineate species in Copelatus. The latter is time consuming and requires specific knowledge of the type of character differences associated with species level separation. Yet the complexities of genotypic and morphological diversity in Copelatus illustrated the difficulty of establishing a Linnaean taxonomy for this group, even if standard taxonomic practice was able to recognize discrete species and to delineate their boundaries. It is possible that in many rapidly radiating lineages neither classical morphological nor DNA barcoding approaches would provide a meaningful system for classification. However, the DNA sequence variation in Copelatus itself, and the evolutionary framework derived from these sequences, represent a synthesis of the available data (a ‘DNA taxonomy’) and provide a basis for comparative analysis and biodiversity studies.
2. Material and methods
(a) Copelatus sampling and sequencing
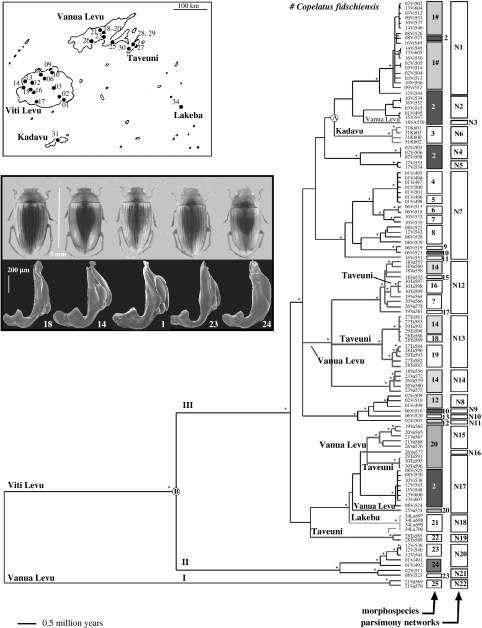
Fiji comprises 332 islands with a total area of ca 18 000 km2 (see inset, figure 1). The archipelago is dominated by two main islands, Viti Levu (10 400 km2) and Vanua Levu (5587 km2). Geological ages range from 25.0 Myr ago for Viti Levu to 3.0–0.7 Myr ago for Taveuni (Nunn 1998; P. D. Nunn 2004, personal communication). Specimens were collected from 25 localities on five islands of the Fijian archipelago (figure 1) in November and December 2003. One or more specimens were chosen from each locality in order to include as many morphologically distinguishable individuals per site as possible. Specimens selected were mostly males, as their genital structures are an important source of information in traditional taxonomic studies. Non-destructive DNA extraction, PCR and sequencing of mitochondrial rrnL (16S rRNA), cox1 (cytochrome oxidase 1), cob (cytochrome oxidase b) regions and the nuclear histone 3 gene were conducted using published methods (Balke et al. 2004). Sequences were aligned by eye to account for minimal length variation in the rrnL. New sequences have been submitted to EMBL under accession numbers AJ 848802–AJ 849303, while most sequences for 18 out-group species of Copelatus from Australia, New Guinea, SE Asia and the Comoro Islands were taken from Balke et al. (2004).
Figure 1.
Maximum likelihood tree (see §2) with branch lengths fitted to a molecular clock by penalized likelihood, resulting in the collapse of several nodes. Terminal labels are indicative of islands (Vi, Viti Levu; Va, Vanua Levu; K, Kadavu; Ta, Taveuni; La, Lakeba). The first two digits of terminal labels correspond to the collecting locality, as given in the inset map of collecting localities. Branches are labelled for lineages found on the three smaller islands. An asterisk (*) indicates bootstrap support greater than 90% in a parsimony analysis. The first column to the right of terminal labels indicates morphological group membership; polyphyletic morphospecies are indicated by shading. The second column indicates statistical parsimony (mtDNA) network membership. Upper inset: map of the Fiji archipelago, depicting the 25 sample localities throughout the five islands studied. Lower inset: dorsal view (above) and SEM of male genital morphology (below) for five representative morphospecies of Fijian Copelatus; numbers correspond to morphospecies designations in the first column to the right of terminal labels.
Following DNA extraction, beetles were dissected, dry-mounted and examined for morphological differences under a stereomicroscope. Morphological characters used to distinguish morphospecies were those traditionally used in taxonomic studies of the group, such as body size and form, colouration, surface sculpture and male genital structure. In particular, the use of male genital morphology for distinguishing otherwise similar (cryptic) species was pioneered by Sharp (1882) and penis structure continues to be a primary means of distinguishing between similar Dytiscidae species (Biström 1997; Miller 2002; Fery 2003). Individuals were sorted into morphological types independently by author M. Balke and by G. Wewalka (Vienna). Both analyses resulted in the same conclusions about morphospecies designation. Morphospecies 1 (see §3) likely refers to C. fidschiensis.
(b) Phylogenetic analysis
Parsimony analysis was conducted in PAUP* 4.0b10 (Swofford 2002) under equal weight of all characters and using the combined matrix of four gene regions. There was minimal length variation in rrnL (481–485 bp) and we examined the effect of gaps on parsimony searches by comparing trees generated using gaps as a fifth character state, as missing data and recoded as binary characters using GapCoder (Young & Healy 2003). For each search, we conducted 1000 random addition replicates, with TBR branch swapping and keeping ≤100 trees per replicate. There was no significant difference in tree likelihood for the three gap settings using a Shimodaira–Hasegawa (SH) test with 1000 Rell bootstrap approximations (Shimodaira & Hasegawa 1999) in PAUP* (all p>0.45) and further analysis was made using gaps as a fifth character. All shortest trees were compared, in order to identify the topology of highest likelihood under a GTR+I+G model (selected in Modeltest 3.06; Posada & Crandall 1998) using the SH test. This optimal tree was further subjected to a 24 h maximum likelihood search in PAUP*, resulting in a small improvement of likelihood scores. Removal of H3 data in order to calculate mitochondrial DNA (mtDNA) branch lengths and estimate divergence times (below) had no effect on topology.
Two methods for grouping of sequences were employed. Multi-dimensional scaling was implemented in Statistica 6.0, using uncorrected p-distances calculated with Mega v. 2.1 (Kumar et al. 2001). Gaps and missing data were excluded from each pairwise calculation. Haplotype networks based on statistical parsimony (Templeton et al. 1992) were generated using Tcs 1.13 (Clement et al. 2000) using only the mtDNA partitions. Statistical networks subdivide the variation based on the level of homoplasy within the data themselves, i.e. distinguish between long (homoplastic) and short (non-homoplastic) branches. This provides a relative measure of divergence within a given dataset, rather than a pre-determined phenetic cut-off value. Uncorrected p-distances were calculated within and among networks using Mega as above.
(c) Estimation of divergence times
The evolutionary time frame of the Fijian radiation was estimated with likelihood branch lengths, based on the mtDNA data under the selected GTR+I+G model. Absolute ages of nodes were estimated by fitting branch lengths using penalized likelihood (PL) with optimal smoothing estimated by cross-validation with the r8s program (Sanderson 2003). We tested four smoothing values (1, 10, 100 and 1000). Sampling error due to stochastic rate changes may affect branch length estimates and hence age estimates. This type of error was assessed by estimating confidence intervals for node ages based on 100 bootstrap replicates of the primary sequence data and calculating branch length on each of the bootstrap data sets in PAUP*. Error assessment was conducted using the original tree topology and parameters estimated in the likelihood search (Baldwin & Sanderson 1998). Finally, branch lengths were made ultrametric using PL and the optimal smoothing value, and age confidences were estimated using the profile command in r8s.
3. Results
(a) DNA diversity, phylogeny and geographic distribution
Sequencing of three mtDNA gene regions resulted in 112 different haplotypes in Fijian individuals (n=118), with 81 haplotypes for cob, 80 for cox1, 29 for rrnL. The parsimony search on the full dataset of Fijian and non-Fijian Copelatus (with Aglymbus as an out-group; 1939 characters, 604 parsimony-informative) resulted in 283 shortest trees of length 2894 (homoplasy index=0.637; retention index=0.739). The strict consensus tree showed Fijian Copelatus were monophyletic and sister to a clade of Australian and New Guinean taxa. The optimal maximum likelihood tree derived from the shortest parsimony trees (see §2) showed identical topology. Many individuals were heterozygous or produced ambiguous reads at one or more H3 bases. These were scored as N for the analysis. As a result, H3 contributed only two parsimony-informative characters (for the Fiji in-group samples) and exclusion of the data had no effect on tree topology. We identified three major Fijian lineages, hereafter referred to as clades I, II and III (figure 1). Clade III contained 109 individuals and included exemplars from all five islands sampled. We identified 26 tip clades corresponding to well-supported nodes in parsimony analyses and characterized by one or more similar individuals (short branches) separated from others by comparatively longer branches. Each tip clade was composed of individuals from only one island, but thirteen (i.e. 50%) included individuals from more than one locality on a given island. The small islands Kadavu and Lakeba were each represented by a single lineage, while Taveuni harboured five distinct lineages from throughout clade III.
(b) Divergence times
Divergence times were estimated using both geological data and sequence divergence. Nodal age calibrations were first obtained by setting the Kadavu–Viti Levu node to 2 Myr ago, based on the geological evidence suggesting Kadavu's age is between 1.5 and 2.5 Myr ago (node A, figure 1). Using this calibration, and based on 100 pseudoreplicates used to estimate node confidence, the basal node of the Fijian Copelatus lineage was inferred to be 9.9 (s.d. 1.8) Myr ago and the base of the most diverse lineage (clade III) was 3.7 (s.d. 0.5) Myr ago. Age calibration using sequence divergence was obtained by applying the widely accepted insect mtDNA molecular clock of 2.3% My−1 (Brower 1994). Mean sequence divergence between individuals from Kadavu and Viti Levu was 2.4% (s.d. 0.3%, uncorrected p-distance), corresponding to 1.04 Myr ago (instead of 2 Myr ago used above). This was slightly more recent than the inferred age of Kadavu (P. D. Nunn 2004, personal communication), and extrapolates to a more recent arrival of Copelatus in Fiji (5.0 Myr ago s.d. 0.8) and diversification of clade III (1.9 Myr ago).
(c) Group delineation based on DNA sequences
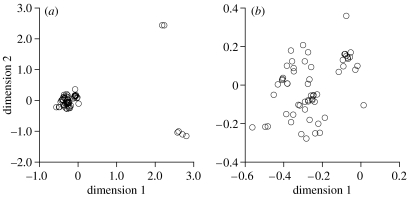
Grouping procedures based on multi-dimensional scaling produced three major clusters (figure 2a), corresponding to clades I, II and III. Mean pairwise divergence among the three major clades was 8.9% (I and II), 9.0% (I and III) and 7.8% (II and III) for cox1 and slightly greater for cob (data not presented). Within clade III, mean cox1 divergence was 3.2% and ranged from 0 to 4.5%; however, a separate analysis failed to show any discrete clusters within this group (figure 2b). Statistical parsimony analysis of mtDNA produced 22 networks separated by a minimum of 17 steps (N1–N22 in figure 1). These corresponded to well-supported monophyletic groups in all but a single case (N16, figure 1). Mean genetic divergence within the 22 mtDNA networks was always less than 1% (table 1). Among networks, divergence ranged from 1.3 to 5.6% (table 1).
Figure 2.
Two-dimensional representation of multi-dimensional scaling analysis using uncorrected p-distances for (a) 65 individuals representative of the entire Fiji in-group and (b) 59 individuals representative of clade III (see figure 1).
Table 1.
Mean, minimum and maximum pairwise genetic divergence (%) between any two sequences within and among the 22 mtDNA haplotype networks (see figure 1). (Divergence was calculated as uncorrected p-distance using Mega v. 2.1 with gaps and missing data treated with pairwise deletion. Minimum values of 0.0 indicate identical haplotypes were present.)
| within networks | among networks | |||||
|---|---|---|---|---|---|---|
| gene | mean (s.d.) | min | max | mean (s.d.) | min | max |
| rrnL | 0.2 (0.2) | 0.0 | 1.0 | 1.3 (0.7) | 0.0 | 3.4 |
| cox1 | 0.5 (0.4) | 0.0 | 1.3 | 4.7 (2.2) | 1.4 | 10.0 |
| cob | 0.9 (0.7) | 0.0 | 2.6 | 5.6 (2.3) | 1.8 | 11.7 |
| combined | 0.4 (0.3) | 0.0 | 0.9 | 3.4 (1.4) | 1.0 | 7.3 |
(d) Groups based on morphology
Male beetles were assigned to 25 distinct morphological groups which, based on the degree of observed differences, would be differentiated as species in current taxonomic practice for Dytiscidae (e.g. Miller 2002; Fery 2003). Three females could not be classified. Nine of the morpho-groups occurred in multiple localities, and two (groups 14 and 20) were found on multiple islands. Each locality contained representatives of one to five sympatric morphological groups, and this number was remarkably similar to the number of parsimony networks present on each island. However, the extent of morphological groups differed from those defined by DNA. Mapped onto the preferred tree, eight morphological groups were paraphyletic (groups 1, 2, 10, 12, 14, 20, 23, 24; figure 1). The morphological groupings were generally more finely subdivided than the parsimony networks, and any one network included between one and eight morphological groups. Only four of the morphologically defined species were completely congruent with the parsimony networks. Three of these were groups from the small islands of Kadavu (N6), Lakeba (N18) and Taveuni (N19). The fourth was represented by a single individual from Viti Levu (N10) not linked to any other in the statistical parsimony analysis. Most incongruence involved closely related networks, and occurred within, rather than among, islands. Only in two cases were individuals of the same morphological group found in widely divergent networks (morphological groups 2 and 14 with mtDNA grouped in clade I and II).
4. Discussion
The Fijian Copelatus exemplify a common situation where the lack of basic information about species identity and distribution hampers our understanding of biological diversity and consequently any efforts for conservation (Abell 2002). We show that DNA-based inventories can provide a rapid assessment for taxonomic classification at the species level and provide an evolutionary framework for the group. The Fijian Copelatus are monophyletic and stem from a single colonization out of the Australian region within the last 10 Myr ago. Lineage diversification occurred predominantly during the recent Pleistocene, particularly in the highly diverse clade III. We encountered a great diversity of haplotypes whose spatial distribution showed complex overlapping ranges. The analysis of mtDNA haplotypes identified clearly separated groups recognizable in the nested clade analysis, but the genetic distance among groups was variable and very low in many instances. Most of these networks included multiple species identified in the morphological analysis, but their extent was frequently incongruent with the DNA-based groupings.
The findings raise questions about the application of DNA barcodes and Linnaean names in ‘taxonomically complex groups’ (Ennos et al. 2005), such as rapidly diversifying island radiations. Least problematic were populations from the small islands of Kadavu and Lakeba, where unique sets of genotypes were narrowly restricted geographically and coincided with the morphological groups. These are distinct species by the criteria of diagnosability (Cracraft 1983) or exclusivity (Wiens & Penkrot 2002), confirming that recognizable species did originate during the evolutionary time frame under investigation here. However, assemblages from all other localities were highly polyphyletic at the scale of islands and at the scale of sampling localities within islands. Twenty of the 22 networks had fully or partially overlapping distributions with members of up to four different networks found at a particular locality. This suggests either that populations on the large islands constitute a single, if highly diverse species, or that multiple sympatric (syntopic) species with various degrees of genetic differentiation coexist in most localities.
The recognition of different morphological groups, established on criteria of phenotypic variation without consideration of their geographic distribution, supports the multiple species scenario. The spatial arrangement of the 25 morphological groups was complex, consisting of sympatric groups that were widespread within an island, but rarely found on multiple islands, confirming wide movement on islands and more restricted movement among islands in agreement with the DNA data. The incongruence of genetic and morphological boundaries suggests a variety of processes are involved in generating the complex patterns. On the one hand, introgression and persistence of ancient polymorphisms is common in island radiations (Gillespie & Roderick 2002; Shaw 2002). Alternatively, morphological convergence of distant genetic groups (Gillespie 2004) also may occur through selection (Losos 1992) or a combination of selection and hybridization (Grant et al. 2004). Further research on these few focal groups would help us to distinguish from among these hypotheses.
The challenge for establishing a formal taxonomic system, therefore, is to capture the complex pattern of variation in DNA, and to reconcile the discrepancies between genetics and morphology. DNA barcoding studies generally find clean separation of species whose haplotype divergences was at least ten times greater than intraspecific distances (Hebert et al. 2003b, 2004), but this was not the case here. Multi-dimensional scaling to identify graphically the discontinuities in sequence space (Hebert et al. 2003a) revealed only the three deeply divergent lineages of the phylogenetic tree, an oversimplification of genotype distribution and diversity. Statistical parsimony analysis provided another means of grouping the sequence variation without the need for defining cohesive populations a priori (Templeton 2001). Previous studies have broadly equated haplotype networks with distinct species in cases where a taxonomic classification and information on geographical isolation had been established independently (Wiens & Penkrot 2002; Morando et al. 2003; Wilder & Hollocher 2003). The finding that the small islands harboured separated networks congruent with morphology is significant in this regard, as species status is supported by multiple sources of evidence (DeSalle et al. 2005; Page et al. 2005).
However, in all other localities the extent of networks and morphological groups did not coincide. Traditional morphological analysis would have accepted the existence of well-defined groups, focusing on traits perhaps under (sexual) selection, but naming them would have formalized at best a partial reality and limited the evolutionary understanding of the lineage. The sequencing approach provided an extensive summary of the evolutionary history and present-day distribution of Fijian Copelatus, demonstrating the complexity of groupings that is also confirmed from the incongruences with the morphological data. However, assigning Linnean names to the DNA-based groups (such as the networks of the statistical parsimony analysis) would be equally inappropriate because of their unclear species status and apparent propensity for gene exchange.
One purpose of a taxonomic system is to provide a point of reference for collecting ‘collateral’ information (Janzen 2004) that can be accumulated under a particular name and made accessible to comparative biology. mtDNA surveys of the kind shown here can assume a similar function, whereby the DNA sequences themselves constitute a system for grouping and communication. This ‘DNA taxonomy’, i.e. the evolutionary framework derived from the sequence information and an analysis of grouping, is the reference point for accumulating ecological, geographical, morphological and other data. This approach can be used where Linnean species names are not (yet) available, or, as in the Fijian Copelatus, where meaningful natural groupings cannot easily be defined from the patterns of variation. DNA taxonomy relies on algorithmic grouping procedures, such as statistical parsimony analysis (Templeton et al. 1992), which cluster closely related sequences and separate them from others. These clusters may be labelled for easy recognition, and can be assessed in a phylogenetic context if the sequences are sufficiently informative (figure 1). The DNA taxonomy of a lineage is a reference system, which is specific to particular (sets of) DNA markers. This provides a new approach to the exploration of taxonomic questions whereby a DNA-based profile is established for entire assemblages without prior knowledge of species membership. The procedure removes the need for specialist morphological analysis (which can be added at a later stage where desired; Tautz et al. 2003). Rather than signalling an end to traditional taxonomy (Seberg et al. 2003), DNA profiling of lineages and natural communities could make the great task of global species classification achievable where standard taxonomic methods are inadequate or simply too time consuming.
Acknowledgments
Bula and Vinaka vakalevu to the many people in Fiji who granted us permission to work on their land and to G. Wewalka and K. Koto for their company in the field. Vinaka to David and Linda Olson, Moala, Alex and especially Kini Koto of the Wildlife Conservation Society South Pacific Program, as well as Prof. G. Wewalka for invaluable support during the entire project. Samples were collected under the Wildlife Conservation Society South Pacific Program's collecting permit. We thank Prof. P. Nunn (USP Suva) for information on island ages and M. Junker (Munich) for assistance with SEM. Two anonymous referees provided constructive comments. Research was funded by the Deutsche Forschungsgemeinschaft (BA 2152/1–2) and The Linnean Society of London. M.T.M. was supported by a Swiss National Science Foundation postdoctoral fellowship (SNF 68592) and BBSRC grant BBS/B/04358. M.B. was a Marie Curie Postdoctoral Fellow (HPMF-CT-2001-01365). A curated collection of vouchers will be repatriated to fulfil the obligations of the Convention on Biological Diversity.
Footnotes
Present address: Zoologische Staatssammlung, Münchhausenstrasse 21, 81247 Munich, Germany.
Present address: Universitat Pompeu Fabra, Facultat de Ciencies de la Salut i la Vida, Unitat de Biologia Evolutiva, Dr Aiguader 80, 08003 Barcelona, Spain.
References
- Abell R. Conservation biology for the biodiversity crisis: a freshwater follow-up. Conserv. Biol. 2002;16:1435–1437. doi: 10.1046/j.1523-1739.2002.01612.x. 10.1046/j.1523-1739.2002.01532.x [DOI] [PubMed] [Google Scholar]
- Baldwin B.G, Sanderson M.J. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proc. Natl Acad. Sci. USA. 1998;95:9402–9406. doi: 10.1073/pnas.95.16.9402. 10.1073/pnas.95.16.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balke M, Ribera I, Vogler A.P. MtDNA phylogeny and biogeography of Copelatinae, a highly diverse group of tropical diving beetles (Dytiscidae) Mol. Phylogenet. Evol. 2004;32:866–880. doi: 10.1016/j.ympev.2004.03.014. 10.1016/j.ympev.2004.03.014 [DOI] [PubMed] [Google Scholar]
- Biström O. Taxonomic revision of the genus Hydrovatus Motschulsky (Coleoptera, Dytiscidae) Entomol. Basiliensia. 1997;19:57–584. [Google Scholar]
- Blaxter M.L. The promise of a DNA taxonomy. Phil. Trans. R. Soc. B. 2004;359:669–679. doi: 10.1098/rstb.2003.1447. 10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl Acad. Sci. USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B, Johnson M.S, Murray J. Clines in the genetic distance between two species of island land snails: how ‘molecular leakage’ mislead us about speciation. Phil. Trans. R. Soc. B. 1996;351:773–784. [Google Scholar]
- Clement M, Posada D, Crandall K.A. Tcs: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Cracraft J. Species concept and speciation analysis. Curr. Ornithol. 1983;1:159–187. [Google Scholar]
- DeSalle R, Egan M.G, Siddall M. The unholy trinity; taxonomy, species delimitation and DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1905–1916. doi: 10.1098/rstb.2005.1722. 10.1098/rstb.2005.1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirzo R, Raven P.H. Global state of biodiversity and loss. Annu. Rev. Environ. Resour. 2003;28:137–167. 10.1146/annurev.energy.28.050302.105532 [Google Scholar]
- Ennos A.E, French G.C, Hollingsworth P.M. Conserving taxonomic complexity. Trends Ecol. Evol. 2005;20:164–168. doi: 10.1016/j.tree.2005.01.012. 10.1016/j.tree.2005.01.012 [DOI] [PubMed] [Google Scholar]
- Fery H. Dytiscidae: V. Taxonomic and distributional notes on Hygrotus Stephens, with emphasis on the Chinese fauna and a key to the Palearctic species (Coleoptera) In: Jäch M.A, Ji L, editors. Water beetles of China. vol. 3. Zoologisch-Botanische Gesellschaft in Österreich and Wiener Coleopterologenverein (Vienna); Wien: 2003. pp. 133–193. [Google Scholar]
- Gillespie R. Community assembly through adaptive radiation in Hawaiian spiders. Science. 2004;303:356–359. doi: 10.1126/science.1091875. 10.1126/science.1091875 [DOI] [PubMed] [Google Scholar]
- Gillespie R.G, Roderick G.K. Arthopods on islands: colonization, speciation and conservation. Annu. Rev. Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. 10.1146/annurev.ento.47.091201.145244 [DOI] [PubMed] [Google Scholar]
- Grant P.R, Grant B.R, Markert J.A, Keller L.F, Petren K. Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution. 2004;58:1588–1599. doi: 10.1111/j.0014-3820.2004.tb01738.x. [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, DeWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, DeWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003b;270(Suppl. 1):S96–S99. doi: 10.1098/rsbl.2003.0025. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:1657–1663. doi: 10.1371/journal.pbio.0020312. 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen D.H. Now is the time. Phil. Trans. R. Soc. B. 2004;359:731–732. doi: 10.1098/rstb.2003.1444. 10.1098/rstb.2003.1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy D.A, Conn J.E. Molecular and morphological phylogenetic analysis of an insular radiation in pacific black flies (Simulium) Syst. Biol. 2001;50:18–38. 10.1080/106351501750107431 [PubMed] [Google Scholar]
- Keast A, Miller S. SPB Academic Publishing; Amsterdam: 1997. The origin and evolution of Pacific island biotas, New Guinea to Eastern Polynesia: patterns and processes. [Google Scholar]
- Kumar S, Tamura K, Jakobsen I.B, Nei M. Mega2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. 10.1093/bioinformatics/17.12.1244 [DOI] [PubMed] [Google Scholar]
- Losos J.B. The evolution of convergent structure in Caribbean Anolis communities. Syst. Biol. 1992;41:403–420. [Google Scholar]
- Mayr E. Harvard University Press; Boston, MA: 1963. Animal species and evolution. [Google Scholar]
- Miller K.B. Revision of the genus Eretes Laporte, 1833 (Coleoptera: Dytiscidae: Dytiscinae: Eretini) Aquat. Insects. 2002;24:247–272. 10.1076/aqin.24.4.247.8238 [Google Scholar]
- Monaghan M.T, Balke M, Gregory T.R, Vogler A. DNA-based species delineation in tropical beetles using nuclear and mitochondrial markers. Phil. Trans. R. Soc. B. 2005;360:1925–1933. doi: 10.1098/rstb.2005.1724. 10.1098/rstb.2005.1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morando M, Avila L.J, Sites J.W. Sampling strategies for delimiting species: genes, individuals and populations in the Liolaemus elongates-kriegi complex (Squamata: Liolaemidae) in Andean–Patagonian South America. Syst. Biol. 2003;52:159–185. doi: 10.1080/10635150390192717. [DOI] [PubMed] [Google Scholar]
- Moritz C, Cicero C. DNA barcoding: promise and pitfalls. PLoS Biol. 2004;2:1529–1531. doi: 10.1371/journal.pbio.0020354. 10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn P.D. Institute of Pacific Studies, The University of the South Pacific; Suva: 1998. Pacific island landscapes. [Google Scholar]
- Page T.J, Choy S.C, Hughes J.M. The taxonomic feedback loop: symbiosis of morphology and molecules. Biol. Lett. 2005;1:139–142. doi: 10.1098/rsbl.2005.0298. 10.1098/rsbl.2005.0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulian R. Societe nouvelle des editions boubee; Paris: 1998. Les insectes de Tahiti. [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. 10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Proudlove G, Wood P.J. The blind leading the blind: cryptic subterranean species and DNA taxonomy. Trends Ecol. Evol. 2003;18:272–273. 10.1016/S0169-5347(03)00095-8 [Google Scholar]
- Sanderson M.J. r8s: inferring absolute rates of molecular evolution, divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. 10.1093/bioinformatics/19.2.301 [DOI] [PubMed] [Google Scholar]
- Seberg O. The future of systematics: assembling the tree of life. The Systematist. 2004;22:2–8. [Google Scholar]
- Seberg O, Humphries C.J, Knapp S, Stevenson D.W, Petersen G, Scharff N, Andersen N.M. Shortcuts in systematics? A commentary on DNA-based taxonomy. Trends Ecol. Evol. 2003;18:63–65. 10.1016/S0169-5347(02)00059-9 [Google Scholar]
- Sharp D. On aquatic carnivorous Coleoptera or Dytiscidae. Scient. Trans. R. Dubl. Soc. 1882;2:179–1003. [Google Scholar]
- Shaw K.L. Conflict between nuclear and mitochondrial DNA phylogenies of a recent species radiation: what mtDNA reveals and conceals about modes of speciation in Hawaiian crickets. Proc. Natl Acad. Sci. USA. 2002;99:16 122–16 127. doi: 10.1073/pnas.242585899. 10.1073/pnas.242585899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2002. PAUP*: Phylogenetic analysis using parsimony. Version 4.0b. [Google Scholar]
- Tautz D, Arctander P, Minelli A, Thomas R.H, Vogler A.P. A plea for DNA taxonomy. Trends Ecol. Evol. 2003;18:70–74. 10.1016/S0169-5347(02)00041-1 [Google Scholar]
- Templeton A.R. Using phylogeographic analyses of gene trees to test species status and processes. Mol. Ecol. 2001;10:779–791. doi: 10.1046/j.1365-294x.2001.01199.x. 10.1046/j.1365-294x.2001.01199.x [DOI] [PubMed] [Google Scholar]
- Templeton A.R, Crandall K.A, Sing C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and sequencing data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens J.J, Penkrot T.A. Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus) Syst. Biol. 2002;51:69–91. doi: 10.1080/106351502753475880. 10.1080/106351502753475880 [DOI] [PubMed] [Google Scholar]
- Wilder J.A, Hollocher H. Recent radiation of endemic Caribbean Drosophila of the dunni subgroup inferred from multilocus DNA sequence variation. Evolution. 2003;57:2566–2579. doi: 10.1111/j.0014-3820.2003.tb01500.x. [DOI] [PubMed] [Google Scholar]
- Will K.W, Rubinoff D. Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics. 2004;20:47–55. doi: 10.1111/j.1096-0031.2003.00008.x. 10.1111/j.1096-0031.2003.00008.x [DOI] [PubMed] [Google Scholar]
- Wilson E.O. The encyclopedia of life. Trends Ecol. Evol. 2003;18:77–80. 10.1016/S0169-5347(02)00040-X [Google Scholar]
- Young N.D, Healy J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinform. 2003;4:6. doi: 10.1186/1471-2105-4-6. 10.1186/1471-2105-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A. Neuer Beitrag zur Kenntnis der Schwimmkäfer. Wien. Entomol. Zeit. 1928;44:165–187. [Google Scholar]
- Zink R.M. The geography of mitochondrial DNA variation in two sympatric sparrows. Evolution. 1991;45:329–339. doi: 10.1111/j.1558-5646.1991.tb04407.x. [DOI] [PubMed] [Google Scholar]