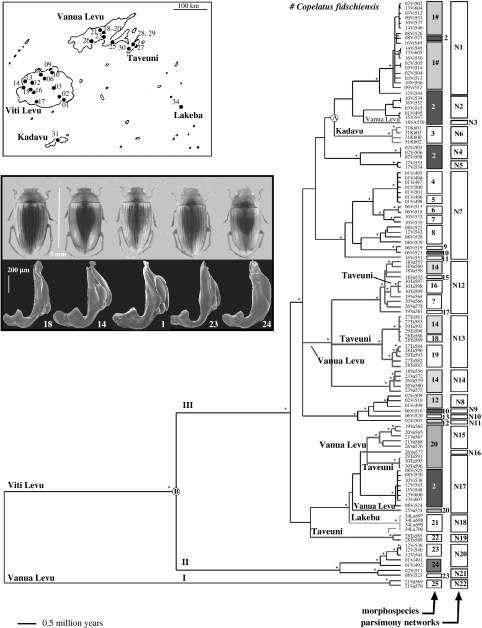
Figure 1.
Maximum likelihood tree (see §2) with branch lengths fitted to a molecular clock by penalized likelihood, resulting in the collapse of several nodes. Terminal labels are indicative of islands (Vi, Viti Levu; Va, Vanua Levu; K, Kadavu; Ta, Taveuni; La, Lakeba). The first two digits of terminal labels correspond to the collecting locality, as given in the inset map of collecting localities. Branches are labelled for lineages found on the three smaller islands. An asterisk (*) indicates bootstrap support greater than 90% in a parsimony analysis. The first column to the right of terminal labels indicates morphological group membership; polyphyletic morphospecies are indicated by shading. The second column indicates statistical parsimony (mtDNA) network membership. Upper inset: map of the Fiji archipelago, depicting the 25 sample localities throughout the five islands studied. Lower inset: dorsal view (above) and SEM of male genital morphology (below) for five representative morphospecies of Fijian Copelatus; numbers correspond to morphospecies designations in the first column to the right of terminal labels.