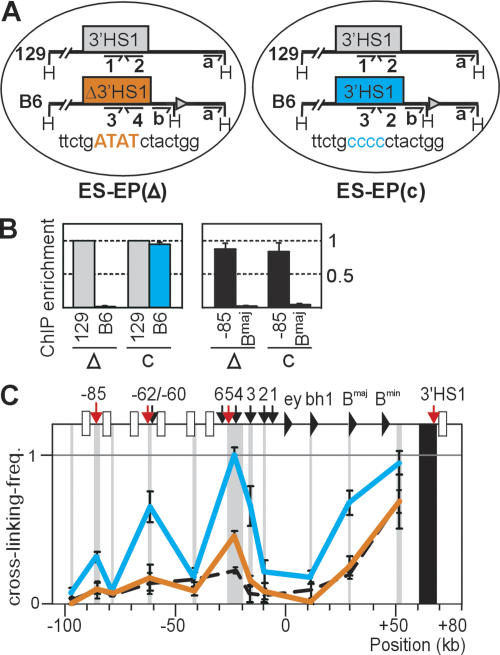
Figure 2.
Targeted nucleotide changes in 3′HS1 disrupt CTCF binding and reduce frequency of long-range 3′HS1 interactions. (A) Erythroid progenitor cell lines derived in vitro from ES cells. ESEP(Δ3′HS1) harbors four targeted nucleotide changes in the core CTCF-binding site of 3′HS1 on the B6 allele (orange). ES-EP(c) contains wild-type 3′HS1 on the B6 allele (blue). The nontargeted, intact 129 allele is in gray. For ChIP, each 3′HS1 CTCF-binding site can be analyzed with a unique primer pair (#1–4). An extra HindIII site targeted downstream from 3′HS1 allows exclusive 3C analysis of B6 allele (with 3C-primer b). (B) ChIP on undifferentiated ES-EP cell lines, with antibody against CTCF. (Left) 3′HS1 alleles in the two ES-EP lines (Δ and c). (Right) Positive (HS-85) and negative (βmajor) controls. (C) 3C analysis with primer b (see above). Note that interaction frequencies with mutated 3′HS1 (orange) are reduced compared with wild-type 3′HS1 (blue). (Black dashed line) 3′HS1 interactions in fetal brain, analyzed with primer a and plotted for comparison.