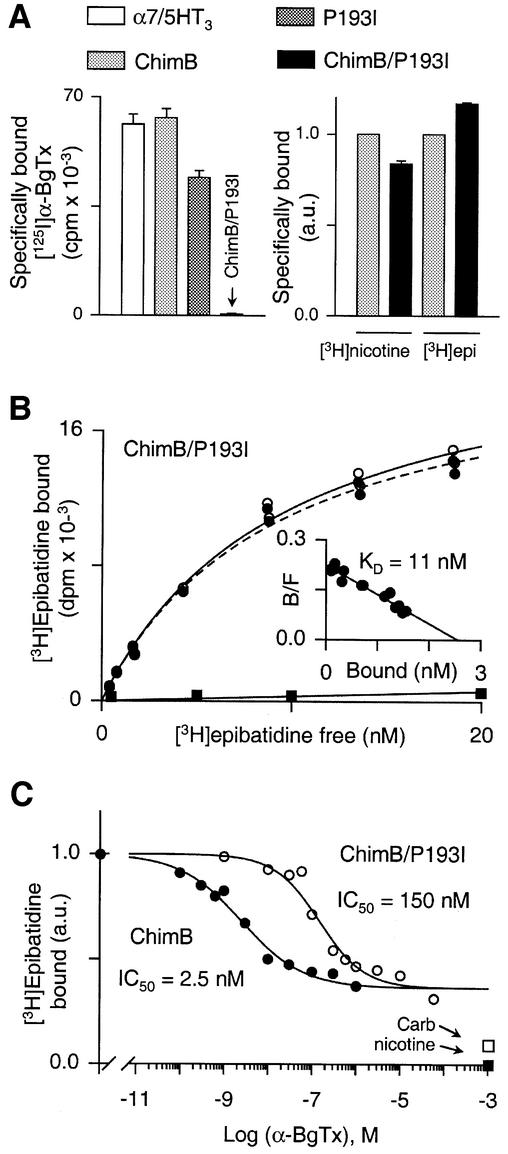
Fig. 2. (A) Left panel: representative experiment of [125I]α-BgTx binding to α7/5HT3 receptor (white bar), ChimB (light gray bar), P193I (dark gray bar) and ChimB/P193I (black bar). Membrane fragments of each construction were incubated with 5 nM [125I]α-BgTx for 2 h and samples were filtered and counted. Values indicated represent specific [125I]α-BgTx binding (non-specific binding was determined in the presence of 1 mM nicotine). Data are means ± SE of triplicates. Right panel: representative experiment of specific (non-specific binding was determined in the presence of 1 mM nicotine) [3H]nicotine and [3H]epibatidine binding on ChimB (light gray bar) and ChimB/P193I (black bar). Membrane fragments were incubated with 100 nM [3H]nicotine or 10 nM [3H]epibatidine until equilibrium. Samples were filtered and counted. Data were normalized (a.u., arbitrary units) to ChimB values and are means ± SE of triplicates. (B) Representative experiment of [3H]epibatidine binding to the double mutant ChimB/P193I. Membrane fragments were incubated with various concentrations of [3H]epibatidine (0.5–20 nM) in the presence (filled squares) or absence (open circles) of 1 mM nicotine until equilibrium and samples were filtered and counted. Specific [3H]epibatidine is also indicated (filled circles). The inset shows a Scatchard plot of specific binding of [3H]epibatidine. Data were fitted to non-linear and linear least-squares analysis for the saturation and Scatchard plots, respectively, according to a single-site model. In this experiment, KD = 11 ± 1 nM and Bmax = 2.56 ± 0.07 nM. (C) Representative competition for [3H]epibatidine binding sites by α-BgTx for ChimB (filled circles) or ChimB/P193I (open circles) microchimeras. Membrane fragments were first incubated with various concentrations of α-BgTx (as indicated) until equilibrium, 10 nM [3H]epibatidine was then added for 1 h and samples were filtered and counted. Data were normalized to values determined in the absence of α-BgTx. Data points are means of duplicates of a representative experiment. Specific [3H]epibatidine binding was determined in the presence of 1 mM nicotine (filled square). Also indicated is [3H]epibatidine binding in the presence of 1 mM carbamylcholine (open square) for both mutants. Data were fitted to non-linear least-squares analysis according to a single-site inhibition model. Note that in this representative experiment, α-BgTx displaced at most 60% of bound [3H]epibatidine, whereas nicotine or carbamycholine displayed almost 100 and 91% of bound [3H]epibatidine, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.