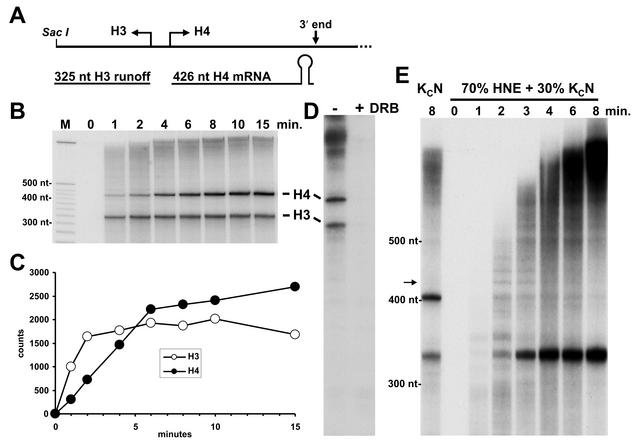
FIG. 1.
Drosophila histone RNAs are transcribed and rapidly processed in nuclear extracts from Drosophila cells. (A) Transcribed region of the Dm3000 plasmid showing H3 runoff transcript and H4 processed mRNA. (B) Pulse-chase assay using Dm3000 template and Kc cell nuclear extract. RNA was isolated at the indicated times and analyzed on a 6% denaturing gel followed by autoradiography. M, DNA markers of indicated sizes. H3 runoff and H4 mRNA are indicated. (C) Quantitation of H3 and H4 transcripts. Transcripts in the dried gel were quantitated by using a Packard InstantImager and the relative counts plotted. (D) Continuous-labeling assay. The Dm3000 template was transcribed in Kc cell nuclear extract in the absence or presence of 50 μM DRB, and the resulting transcripts were analyzed on a 6% denaturing gel. (E) Inhibition of RNA processing in a mixture of HeLa and Kc cell nuclear extracts. Reactions similar to those shown in panel B were carried out except that 70% HeLa and 30% Kc cell nuclear extract were used. Samples were collected at the indicated time points. For comparison purposes, one reaction was carried out for 8 min in the presence of only Kc cell nuclear extract. The transcripts were analyzed in a 6% denaturing gel as described above except that the gel was run longer to better separate long transcripts.