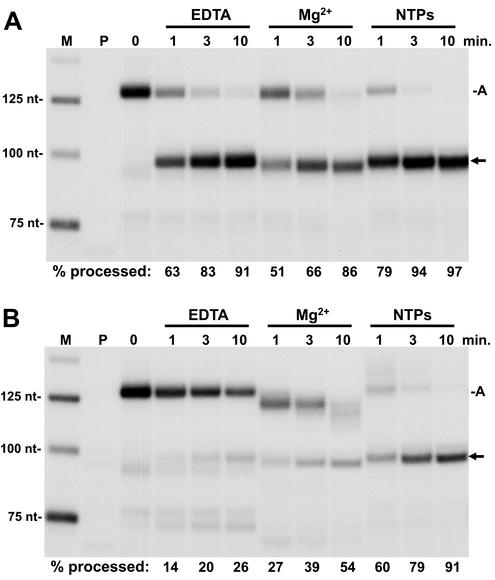
FIG. 4.
Comparison of processing of free RNA to RNA in a transcription complex. Early elongation complexes generated during initiation with a pulse-labeling on the H4 minigene template (lane marked P) were isolated and washed with a high salt concentration and Sarkosyl as described in Materials and Methods. These complexes were chased for 8 min, reisolated, and phenol extracted to obtain free RNA or used directly as elongation complexes. (A) Processing of free RNA. Free RNA containing predominantly the arrested transcript was subjected to processing by nuclear extract for the indicated times with EDTA, Mg, or NTPs with Mg added. (B) Processing of RNA in elongation complexes. Reactions were identical to those in panel A except that the RNA was in a transcription complex. For both panels A and B, RNA was isolated at the indicated times and analyzed on a 6.7% denaturing gel, followed by autoradiography and quantitation with a Packard InstantImager. The percentage of total RNA processed is given under each lane. A, arrested transcript; arrow, processed H4 minigene RNA.