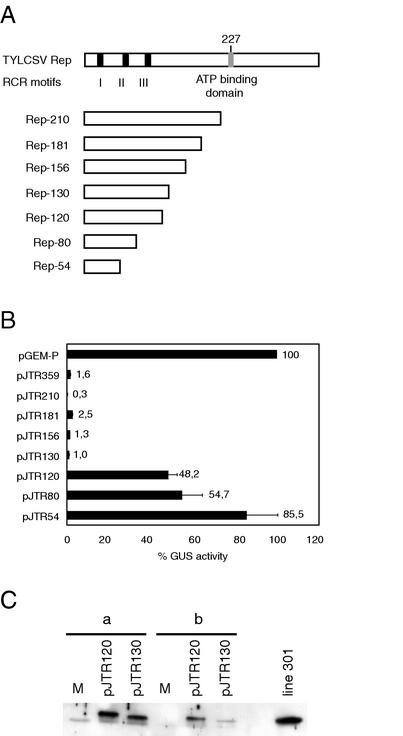
FIG. 1.
Transcriptional repression activity of TYLCSV Rep mutants. (A) Scheme of TYLCSV Rep wild-type and mutant proteins. Solid bars indicate the conserved motifs characteristic of RCR proteins (I, II, and III); a gray bar indicates the ATP binding domain. Rep mutants are C-terminal deletions; the number of retained amino acids is indicated on the left. (B) GUS activity in N. benthamiana protoplasts cotransfected with pTOM202 together with the plasmid indicated on the left. GUS activity in protoplasts cotransfected with pTOM202 and pGEM-P was taken as 100%. Each value is the average of three cotransfections of three independent experiments. Error bars indicate the standard error of the mean. (C) Western blot of protein extracts of N. benthamiana protoplasts transfected with the plasmids indicated above each lane or mock transfected (M). Lane groups a and b represent two independent transfection experiments. Protein extracts from transgenic N. benthamiana protoplasts of line 301 expressing Rep-130 are on the right. The primary antibody was a rabbit polyclonal anti-TYLCSV Rep antibody.