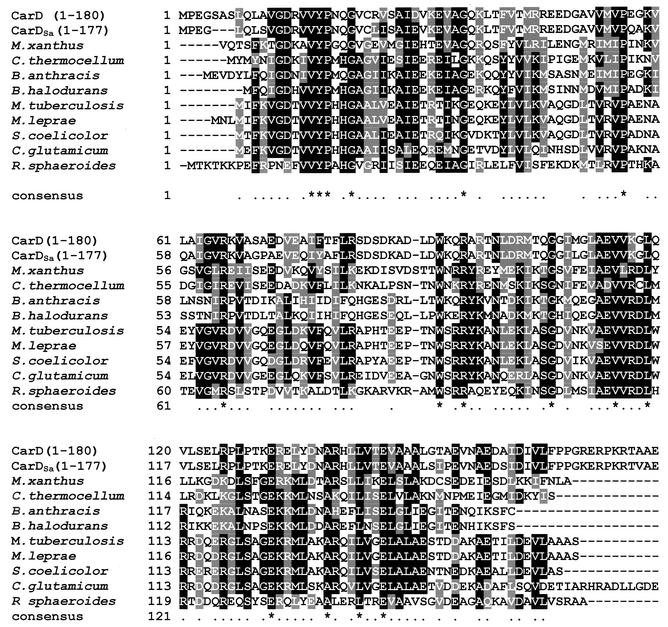
FIG. 6.
Sequence alignment of the 180-residue N-terminal regions of CarDSa, CarD, and the nine bacterial proteins with the highest similarity drawn from the microbial genome sequence database. Accession numbers and total polypeptide lengths are, respectively, JC6146 and 316 residues for M. xanthus CarD; no accession number presently available and 164 residues for M. xanthus; ZP_00061994 and 160 residues for Clostridium thermocellum; NP_657551 and 158 residues for Bacillus anthracis; NP_244803 and 153 residues for Bacillus halodurans; NP_218100 and 162 residues for M. tuberculosis; NP_301347 and 165 residues for Mycobacterium leprae; NP_628406 and 160 residues for Streptomyces coelicolor; NP_601860 and 198 residues for C. glutamicum; and ZP_00005573 and 169 residues for Rhodobacter sphaeroides. The sequence for the 305-residue CarDSa is from this study. Residues are shaded only when they are identical or similar in the majority (≥6) of the 11 aligned sequences described above. Of these, identical residues are shaded black only if they appear in at least six of the sequences. An asterisk in the line corresponding to the consensus indicates identical residues when conserved in all of the aligned sequences.