Abstract
Aspartyl-tRNA synthetase (AspRS) occurs in two types: the discriminating enzyme (D-AspRS) forms only Asp-tRNAAsp, whereas the nondiscriminating enzyme (ND-AspRS) also synthesizes Asp-tRNAAsn, which is a required intermediate for protein synthesis in many organisms. We attempted to expand the tRNA recognition of the discriminating Thermococcus kodakaraensis AspRS to that of a ND-AspRS by in vitro mutagenesis. An alignment of 26 archaeal AspRS proteins revealed two positions (26 and 85 in the T. kodakaraensis sequence) whose amino acid identity changes according to the enzymes' tRNA specificity. In their anticodon-binding domain, D-AspRS proteins contain W26 (or Q26) and K85, compared with H26 and P85 in the ND-AspRSs. T. kodakaraensis AspRS gained the ability to form Asp-tRNAAsn in vitro when the W26H or K85P changes were introduced independently or in combination. In the aminoacylation of tRNAAsn or tRNAAsp transcripts, the mutant enzymes displayed at least a 100- to 500-fold change in tRNA specificity, as judged by the ratio of the kcat/Km values of Asp-tRNAAsp vs. Asp-tRNAAsn formation. That T. kodakaraensis mutant AspRSs mischarge tRNAAsn was also manifested in the higher level (1.7%) of aspartylation of unfractionated Pyrococcus tRNA compared with that achieved by the wild-type enzyme (0.9%). Northern blot analysis of the Asp-tRNA separated by acid/urea gel electrophoresis confirmed the in vitro synthesis of Asp-tRNAAsn. A structure-based model points to a direct interaction of K85 in T. kodakaraensis AspRS with the anticodon nucleotide C36 of tRNAAsp. Thus, a switch between D-AspRS and ND-AspRS enzymes could have evolved with only limited amino acid changes.
Aminoacyl-tRNA synthetases (AARSs) catalyze the formation of aminoacyl-tRNA, the substrate for ribosomal protein synthesis (1). These enzymes possess very high specificity in selecting their cognate amino acid and tRNA substrates, so that misaminoacylation, the charging of the noncognate amino acid to the tRNA, occurs only infrequently (reviewed in ref. 2).
However, there exists a group of misaminoacylating AARSs that are essential for protein synthesis (1). These unusual enzymes, the nondiscriminating aspartyl-tRNA synthetases (ND-AspRSs) and the nondiscriminating glutamyl-tRNA synthetases (ND-GluRSs), are required for Asn-tRNAAsn and Gln-tRNAGln biosynthesis, respectively. The nondiscriminating AspRS (ND-AspRS) charges both tRNAAsp and tRNAAsn with aspartate, whereas the discriminating AspRS (D-AspRS) forms only Asp-tRNAAsp (3, 4). Similarly, a ND-GluRS synthesizes Glu-tRNAGln in addition to Glu-tRNAGlu (e.g., refs. 5 and 6). The misacylated tRNAs are the required substrates for the tRNA-dependent amidotransferases, essential enzymes that carry out Asn-tRNA and Gln-tRNA formation in many organisms and organelles (7–9). Thus, these nondiscriminating synthetases have one cognate amino acid but two “cognate” tRNA substrates. Seryl-tRNA synthetase possesses similar properties as it recognizes both tRNASer and tRNASec species during the process of selenocysteinyl-tRNA formation (10).
Phylogenetic analysis of AspRS sequences reveals the existence in nature of archaeal and bacterial genres of this enzyme (11). The archaeal genre comprises all archaeal AspRS enzymes, as well as one of two types of AspRSs found in the bacteria Thermus thermophilus (12), Deinococcus radiodurans (13), and Clostridium acetobutylicum. Whereas the archaeal AspRS enzymes were thought to be nondiscriminating (12, 14), it is now known that in several archaea (including Thermococcus kodakaraensis), this enzyme is discriminating (15). In contrast to bacterial AspRS proteins, the tRNA specificity of the archaeal enzyme can be predicted from the whole genome content (15): a ND-AspRS always exists in the genome together with the archaeal Asp-tRNAAsn amidotransferase, whereas a D-AspRS accompanies an asparaginyl-tRNA synthetase (AsnRS). D-AspRS enzymes have been well studied, both biochemically and structurally. Crystal structures of AspRSs from diverse organisms exist, including the archaeon T. kodakaraensis (14), Escherichia coli (16), T. thermophilus (17), and yeast (18). Recognition of tRNAAsp by AspRS has been mapped out in detail in bacterial and eukaryotic systems, but not in archaeal systems (19). The major identity elements of bacterial or yeast tRNAAsp include the discriminator base G73 and the anticodon triplet G34, U35, and C36 (20, 21). Crystal structures of AspRS complexed with tRNAAsp from yeast (18) and from E. coli (16) reveal that the tRNA anticodon interacts with the N-terminal (anticodon-binding) domain of the enzyme. The last anticodon base, C36, differentiates tRNAAsp from tRNAAsn and is recognized by the anticodon-binding loop connecting the last two strands of the β-barrel of the class IIb AARSs (16, 18). Despite all these data, there is currently no information on the structural elements in AspRS that determine tRNAAsn selection.
Here we report an in vitro study in which the discriminating T. kodakaraensis AspRS was converted into a nondiscriminating enzyme.
Materials and Methods
General.
Oligonucleotide synthesis and DNA sequencing were performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. [14C]Asp (207 mCi/mmol; 1 Ci = 37 GBq) and [3H]Asp (26 Ci/mmol) were purchased from Amersham Biosciences (Piscataway, NJ) and [14C]Asn (228.4 mCi/mmol) was purchased from Perkin–Elmer Life Sciences (Boston).
Cloning and Purification of the AspRS Proteins.
Cloning and purification of AspRS from T. kodakaraensis, previously known as Pyrococcus kodakaraensis (22), has been described (15). Mutant AspRS genes were generated by overlap-extension PCR and were cloned into pET20b between NdeI and BamHI sites so as to yield native proteins after overexpression. Purification of the mutant AspRSs was the same as for the wild-type enzyme (15). The sequences of the cloned AspRS genes were verified from minipreparations of the cultures from which the AspRSs were purified. The E. coli AsnRS clone was kindly provided by Benfang Ruan (Yale University) and was subcloned into pET28a. The N-terminally His6-tagged protein was purified according to the manufacturer's protocol (Qiagen, Valencia, CA). All enzymes were >95% pure, as judged by SDS/PAGE followed by Coomassie blue staining. Methanothermobacter thermautotrophicus AspRS was prepared as described (15).
Preparation of tRNA and tRNA Transcripts.
Unfractionated Pyrococcus furiosus tRNA and Ferroplasma acidarmanus tRNAAsp and tRNAAsn transcripts were prepared as described (15).
Aminoacylation and Transamidation Assays.
T. kodakaraensis AspRS activity was determined to be optimal at 60°C when the temperature range of 37–70°C was tested. The assay conditions are as follows: 50 mM Mes–KOH, pH 6.0/50 mM KCl/16 mM MgCl2/5 mM DTT (15). The aminoacylation of F. acidarmanus tRNAAsp and tRNAAsn transcripts was carried out in the above buffer containing 150 μM [3H]aspartate (2.6 Ci/mmol), 10 mM ATP, 0.2–19 μM transcript, and 100–150 nM enzyme. Transcripts of F. acidarmanus were used because of the poor chargeability of the more closely related P. furiosus tRNA transcripts (15). For measurement of kinetic parameters, initial velocities of aminoacylation were determined from the average of duplicate sets of data at various tRNA concentrations. The percentage of renatured tRNA that could be aminoacylated was determined for each transcript and varied from 30% to 50% for tRNAAsp and from 50% to 70% for tRNAAsn, respectively. The concentration reported, therefore, reflects the amount of chargeable tRNA. The concentration of aspartate was kept at least 2-fold higher than the Km for the amino acid. The values of kcat and Km were calculated by nonlinear regression fitting of the data to the Michaelis–Menten equation.
Transamidation of Asp-tRNAAsn was carried out as described (15).
Acid/Urea Gel Analysis.
Unfractionated P. furiosus tRNA (20 μM) was aminoacylated with 160 μM unlabeled aspartate and 0.1 μM T. kodakaraensis or M. thermautotrophicus AspRS at 55°C for 30 min as described (15), or with 160 μM unlabeled asparagine and 1 μM E. coli AsnRS at 37°C for 30 min in 50 mM Hepes–KOH, pH 7.0/50 mM KCl/10 mM MgCl2/4 mM ATP/5 mM DTT. Duplicate reactions with the same amount of 14C-labeled amino acid were run in parallel to determine the aminoacylation time course. Aminoacylated and deacylated tRNA samples (4 μg each) were prepared and separated by an acid/urea gel (9.5%) at 4°C for 39 h as described (23). After transfer of the tRNA-containing portion of the gel to a Hybond N+ membrane (Amersham Biosciences), Northern analysis was performed with a 32P-labeled oligonucleotide complementary to bases 11–28 of P. furiosus tRNAAsn in the presence of excess unlabeled oligonucleotide (20 nM) complementary to bases 15–32 of tRNAAsp. The probe is specific for tRNAAsn and does not crossreact with tRNAAsp under the conditions used (data not shown).
Superimposition of AspRS Structures.
Coordinates of crystal structures of T. kodakaraensis AspRS (PDB ID code 1B8A; ref. 14), E. coli AspRS complexed with tRNAAsp and aspartyl-adenylate (PDB ID code 1C0A; ref. 16), and T. thermophilus AspRS1 complexed with E. coli tRNAAsp (PDB ID code 1EFW; ref. 17) were obtained from the Protein Data Bank (www.rcsb.org/pdb). The Cα atoms of 87 residues within the anticodon-binding region of T. kodakaraensis AspRS were superimposed onto those equivalent in E. coli AspRS or T. thermophilus AspRS1 by the least-squares fit method using the program o (24).
Results
Two Amino Acids in the N-terminal Region May Define the tRNA Selection of the Archaeal-Type AspRS.
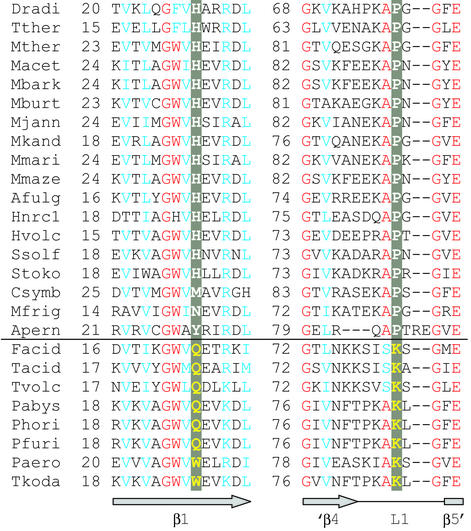
The discriminating and nondiscriminating archaeal-type AspRS enzymes are highly similar to each other. In an alignment of 26 AspRS proteins of predictable tRNA selectivity (15), the highest identity (≈60%) among the two types is between the D-AspRS of the Pyrococcus/Thermococcus group and the ND-AspRS from Archaeoglobus fulgidus or Methanopyrus kandleri. However, the alignment revealed that amino acids at two positions varied systematically between D-AspRS and ND-AspRS (Fig. 1). Both residues are located in the enzyme's N-terminal region. The first amino acid (position 26 in T. kodakaraensis AspRS) is a tryptophan or glutamine in the discriminating enzymes, whereas a histidine is found in most ND-AspRSs. The second amino acid (position 85 in T. kodakaraensis AspRS) is a lysine in the D-AspRS enzymes, whereas it is a proline in the ND-AspRSs. K85 is part of the L1 loop that connects the last two β-strands of the anticodon-binding domain in T. kodakaraensis AspRS (14). The equivalent loop of bacterial and eukaryotic AspRS enzymes interacts with the tRNAAsp anticodon (16, 18). No other significant amino acid differences could be identified in the alignment of the entire AspRS sequence.
Figure 1.
Sequence alignment of part of the N-terminal regions of archaeal-type AspRS proteins. (Upper) ND-AspRSs. (Lower) D-AspRSs. Positions with >90% identity are red, and those with >90% similarity are blue. Amino acids in the two positions that differ between the two groups are highlighted in gray, and shown in white for the ND-AspRSs, and in yellow for the D-AspRSs. (Lower) T. kodakaraensis AspRS secondary structure elements (ref. 14) are indicated; only the C-terminal part of β-strand 4 (′β4) and the N terminus of β-strand 5 (β5′) are shown. Sequences not referenced earlier (ref. 15) are listed with GenBank accession numbers. Afulg, A. fulgidus; Apern, Aeropyrum pernix; Csymb, Cenarchaeum symbiosum (Edward DeLong and Christa Schleper, personal communication); Dradi, Deinococcus radiodurans; Facid, Ferroplasma acidarmanus; Hnrc1, Halobacterium sp. NRC-1; Hvolc, Haloferax volcanii (AB010464); Macet, Methanosarcina acetivorans; Mbark, Methanosarcina barkeri Fusaro (ZP_00078294); Mburt, Methanococcoides burtonii (http://genome.ornl.gov/microbial/mbur/); Mfrig, Methanogenium frigidum (Rick Cavicchioli, personal communication); Mjann, Methanocaldococcus jannaschii; Mkand, Methanopyrus kandleri; Mmari, Methanococcus maripaludis (John A. Leigh, personal communication); Mmaze, Methanosarcina mazei (AAM29770); Mther, Methanothermobacter thermautotrophicus; Pabys, Pyrococcus abyssi; Paero, Pyrobaculum aerophilum; Pfuri, P. furiosus; Phori, Pyrococcus horikoshii; Ssolf, Sulfolobus solfataricus; Stoko, Sulfolobus tokodaii; Tacid, Thermoplasma acidophilum; Tkoda, T. kodakaraensis; Tther, Thermus thermophilus; and Tvolc, Thermoplasma volcanii.
K85 Is Important for the Discriminating Nature of T. kodakaraensis AspRS.
To test the significance of W26 and K85 for tRNA selectivity of T. kodakaraensis AspRS, W26H and K85P mutations were introduced independently, and in combination, into T. kodakaraensis AspRS. Although T. kodakaraensis grows optimally at 95°C (25), in vitro aminoacylation assays were optimal at 60°C because of the instability of Asp-tRNA at higher temperatures. Thus, the kcat values presented in Table 1 are expected to be below physiological values, but comparisons of wild-type and mutant enzymes should show the relative effects of these mutations on the different enzymes.
Table 1.
Kinetic parameters of aspartylation with wild-type and mutant T. kodakaraensis AspRSs
| Enzyme | tRNAAsp
|
tRNAAsn
|
Specificity* | ||||
|---|---|---|---|---|---|---|---|
| kcat, s−1 | Km, μM | kcat/Km, M−1⋅s−1 | kcat, s−1 | Km, μM | kcat/Km, M−1⋅s−1 | ||
| WT | 0.061 ± 0.004 | 1.4 ± 0.2 | 45,000 | ND | ND | <40† | >1,000 |
| W26H | 0.116 ± 0.005 | 4.5 ± 0.5 | 26,000 | 0.026 ± 0.002 | 9.1 ± 1.1 | 2,900 | 9.0 |
| K85P | 0.061 ± 0.003 | 11 ± 1.0 | 5,800 | 0.009 ± 0.001 | 3.1 ± 0.5 | 2,900 | 2.0 |
| W26H/K85P | 0.034 ± 0.002 | 1.2 ± 0.2 | 28,000 | 0.037 ± 0.002 | 6.2 ± 0.7 | 6,000 | 4.7 |
ND, Not determined.
Specificity is the ratio of the kcat/Km values of Asp-tRNAAsp formation over that of Asp-tRNAAsn.
The upper limit of the kcat/Km value for Asp-tRNAAsn formation catalyzed by wild-type T. kodakaraensis AspRS is estimated from the data of Fig. 2.
The K85P mutant enzyme was impaired in its ability to synthesize Asp-tRNAAsp in vitro; it had the same kcat as the wild-type enzyme, but an 8-fold increase in the Km value for tRNAAsp (Table 1). This finding suggests that the conserved lysine in the loop L1 is important for tRNAAsp recognition/binding in the D-AspRS enzymes. In comparison, the W26H enzyme showed a slight increase in both kinetic parameters, resulting in only a 1.5-fold decrease in the overall catalytic efficiency (kcat/Km) for Asp-tRNAAsp synthesis (Table 1). Compared with the wild-type enzyme, the double mutant W26H/K85P had a similar Km for tRNAAsp, but only half the kcat, suggesting a reduced specificity toward tRNAAsp. The simultaneous “change” of both amino acids involved in tRNA recognition may lead to better substrate interaction compared with that seen with the single-mutant enzymes. Although these experiments were performed at suboptimal temperatures and with heterologous transcripts, the Km value for tRNAAsp of the wild-type enzyme (1.4 μM) compares reasonably with that of E. coli AspRS (0.6 μM; ref. 26).
Mutant T. kodakaraensis AspRSs Can Recognize and Charge tRNAAsn.
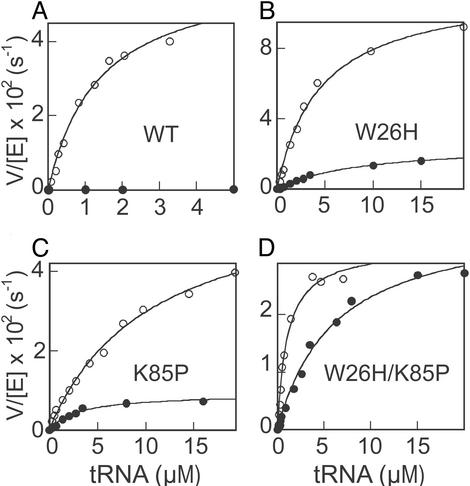
Because all of the mutant enzymes were active, we then determined whether they gained the ability to form Asp-tRNAAsn in vitro. Asp-tRNAAsn synthesis by wild-type T. kodakaraensis AspRS was undetectable at low tRNA concentrations (Fig. 2 and ref. 15). However, by using 5 μM tRNAAsn, a very small amount of product was observed, giving rise to an initial rate of 2 × 10−4 s−1 and a value of 40 M−1⋅s−1 as the upper limit of kcat/Km for Asp-tRNAAsn formation. Compared with the value of 45,000 M−1⋅s−1 for Asp-tRNAAsp formation (Table 1), wild-type T. kodakaraensis AspRS is at least 1,000-fold more specific in selecting tRNAAsp over tRNAAsn (Table 1). In contrast, all three mutant enzymes showed significant levels of Asp-tRNAAsn formation and dramatic decreases (at least 100- to 500-fold) in tRNA discrimination (Table 1 and Fig. 2). The K85P mutant enzyme exhibited the lowest discrimination against tRNAAsn and a 500-fold change in tRNA specificity (Table 1). Therefore, tRNAAsn was aspartylated just 2-fold less efficiently than the cognate tRNAAsp by the K85P enzyme, and this level of nondiscrimination is similar to that of naturally occurring ND-AspRSs (3). In addition, the Km value for tRNAAsn (3.1 μM) of the K85P mutant enzyme was even 3.5 times lower than that for tRNAAsp. The catalytic efficiency of misaminoacylation was comparable between K85P and W26H, but the latter is 9 times more specific for tRNAAsp than for tRNAAsn (Table 1). Of the enzymes tested, W26H/K85P showed the highest activity (kcat/Km) in Asp-tRNAAsn formation (Table 1). All three mutant T. kodakaraensis AspRS enzymes showed a Km value for tRNAAsn ranging from 3 to 9 μM (Table 1).
Figure 2.
tRNA dependence of V/[E] for aminoacylation of tRNAAsp (○) or tRNAAsn (●) by T. kodakaraensis AspRSs. The lines represent hyperbolic fit of the data to the Michaelis–Menten equation (see Table 1 for details).
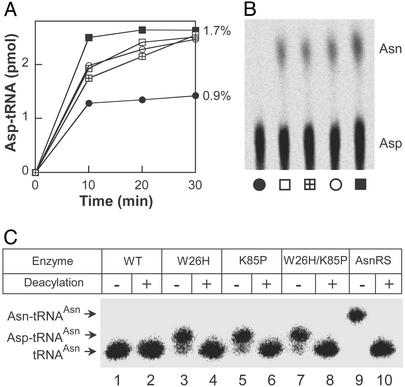
Because the experiments above were performed with unmodified tRNA gene transcripts, we wanted to confirm the tRNA recognition properties of the wild-type and mutant enzymes with mature tRNA from P. furiosus, a close phylogenetic relative of T. kodakaraensis. As shown previously (ref. 15; see also Fig. 3A), the discriminating wild-type T. kodakaraensis AspRS charges unfractionated P. furiosus tRNA to about half the level obtained with the M. thermautotrophicus enzyme that recognizes both tRNAAsp and tRNAAsn. However, the three mutant T. kodakaraensis enzymes charged approximately to the level achieved by the M. thermautotrophicus AspRS (Fig. 3A). This result is consistent with the expectation that the mutant T. kodakaraensis AspRSs can form Asp-tRNAAsn. The presence of this mischarged tRNA was documented (Fig. 3B) by its conversion to Asn-tRNAAsn by purified M. thermautotrophicus Asp-tRNAAsn amidotransferase (15).
Figure 3.
Aspartylation of unfractionated P. furiosus tRNA by T. kodakaraensis AspRSs. (A) Wild-type (●), W26H (□), K85P (⊞), W26H/K85P (○) T. kodakaraensis AspRS, and M. thermautotrophicus AspRS (■). The amount of Asp-tRNA formed, as a percentage of total tRNA, is given. (B) TLC separation of Asn and Asp isolated from aminoacyl-tRNA after transamidation of Asp-tRNAAsn synthesized by wild-type and mutant AspRSs (labeled as in A). The lower amount of Asn (compared with Asp) is due to incomplete transamidation of the heterologous Pyrococcus Asp-tRNAAsn under the conditions used. (C) Northern blot analysis of Pyrococcus tRNA aspartylated by T. kodakaraensis AspRSs or asparaginylated by E. coli AsnRS. The resulting aminoacyl-tRNA was loaded onto an acid/urea gel directly (−) or after deacylation (+). The samples were hybridized with a tRNAAsn-specific oligonucleotide probe. The positions of tRNAAsn, Asp-tRNAAsn, and Asn-tRNAAsn are indicated.
Direct proof of Asp-tRNAAsn formation by the mutant T. kodakaraensis AspRS enzymes was obtained by acid/urea gel analysis (23). Unfractionated P. furiosus tRNA charged with aspartate by purified wild-type and mutant AspRS enzymes was separated by acid/urea gel electrophoresis and then hybridized to a tRNAAsn-specific probe. The W26H, K85P, and double-mutant enzymes, but not wild-type, formed Asp-tRNAAsn (Fig. 3C; compare lanes 3, 5, and 7 with lane 1), which could be deacylated (Fig. 3C; lanes 4, 6, and 8). As expected, Asn-tRNAAsn, made by E. coli AsnRS, moved more slowly than Asp-tRNAAsn (Fig. 3C, lane 9).
K85 of T. kodakaraensis AspRS May Interact with the Anticodon Nucleotide C36 of tRNAAsp.
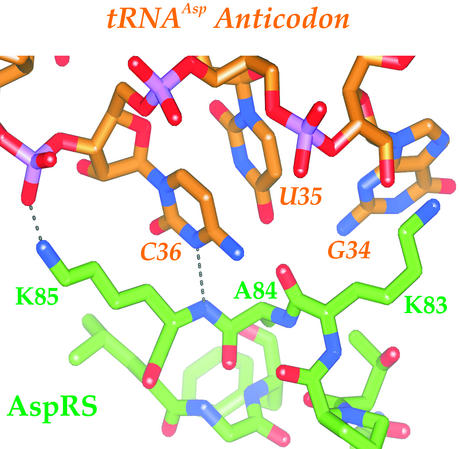
To shed more light on the role of the two amino acids (Fig. 1) in tRNA recognition by an archaeal AspRS, we looked for possible interactions of T. kodakaraensis AspRS with tRNAAsp. Although the crystal structure of this enzyme was determined without tRNA (14), the tRNA complexes of E. coli (16) and T. thermophilus (17) AspRSs are known. These structures are relevant, as it appears that the anticodon-binding domain of T. kodakaraensis AspRS is similar to the corresponding domain in the bacterial counterparts (14). We therefore superimposed the N-terminal domain of T. kodakaraensis AspRS with that of E. coli or T. thermophilus AspRS, both complexed with E. coli tRNAAsp. Similar interactions were observed in both superimpositions, but a lower rms deviation (1.2 Å) was obtained with the E. coli structure (Fig. 4).
Figure 4.
Model of tRNAAsp anticodon recognition by T. kodakaraensis AspRS. Two hydrogen bonds (dotted lines) connect the identified amino acid K85 to the anticodon loop. Oxygen, nitrogen, and phosphorus atoms are shown in red, blue, and purple, respectively. The figure was generated by SPOCK software (27).
Based on this superimposition, K85 in the loop L1 of T. kodakaraensis AspRS interacts directly with tRNAAsp (Fig. 4). Specifically, the peptide backbone amide of K85 donates a hydrogen to the N3 group of C36, the third anticodon base in tRNAAsp. The ɛ-amino group of K85 donates another hydrogen bond to the phosphate backbone of A37 in the tRNA. In contrast, tRNAAsn with U as the third anticodon nucleotide cannot accept this hydrogen bond because, unlike cytidine, uridine is protonated at N3. The superimposition further predicts how the K85P mutation may allow mischarging. As proline is the only amino acid that lacks a backbone hydrogen group, the K85P mutant loses the hydrogen bond with the C36. A K85P mutant also loses the hydrogen bond between the lysine side chain and the A37 phosphate group. Presumably, because of the lack of interactions between the P85 and the tRNA, either cytidine or uridine would fit comfortably at the third anticodon position, when the tRNA is in complex with the AspRS.
W26 of T. kodakaraensis AspRS does not appear to contact the tRNA in this model; however, this residue may affect tRNA recognition indirectly by its vicinity to K29 and D30, amino acids that contact the anticodon loop bases C32 and C38 (16, 17).
Discussion
Changing the tRNA Specificity of an AARS.
Specific tRNA recognition by an AARS relies on a multifaceted interaction of protein and RNA. Studies on tRNA identity made clear that the anticodon (position 34–36), the discriminator base (position 73), and base pairs in the acceptor helix are of primary importance in synthetase recognition (19). As these structurally separated nucleotides are recognized by spatially noncontiguous elements in the synthetase protein, a desired change in tRNA binding by this protein may mandate multiple amino acid changes (28, 29). The task should be easier in the case of the set of discriminating or nondiscriminating AARSs, because the latter enzyme acylates with its cognate amino acid two different acceptor RNA types. These tRNAs must share many identity elements to be accommodated in the enzyme's single tRNA-binding site, which tolerates at least two different anticodons. Part of this task was accomplished when it was shown that a single R358Q mutation endowed the discriminating T. thermophilus GluRS with the ability to recognize tRNAGlu and a tRNAGlu variant containing a tRNAGln anticodon with comparable efficiencies (30). In the case of T. kodakaraensis AspRS, a complete change was effected by the K85P mutation in the enzyme's anticodon-binding domain. Whereas this mutation sufficed to give rise to a ND-AspRS, additional interactions (e.g., position 26) undoubtedly ensure the specificity. The success of our experiments may be because of the high similarity of archaeal tRNAAsp and tRNAAsn sequences. These tRNAs share the discriminator base, the first two anticodon bases, and the first three base pairs of the acceptor arm, regions that contain the identity elements of AsnRS and AspRS (19). However, the ultimate understanding of this dual tRNA recognition will require in vivo proof and crystal structures of a ND-AspRS complexed to the different tRNAs.
Unique Mechanism of tRNAAsx Discrimination by the Archaeal-Type AspRS?
The two amino acids described here (Fig. 1) are not conserved in the bacterial-type AspRS proteins, which also occur in a discriminating (e.g., in E. coli) and a nondiscriminating form (e.g., in Chlamydia trachomatis; ref. 6). This difference may be related to the divergence of the tRNAAsn sequences between the bacterial and archaeal domain. For example, a U1⋅A72 base pair is found in bacterial tRNAAsn, but the corresponding base pair in archaeal tRNAAsn is G1⋅C72. Eukaryotic AspRS proteins show a poor alignment with their archaeal counterparts in the region pictured in Fig. 1, yet there is no need for the existence of a eukaryotic ND-AspRS. Therefore, the evolution of Asx-tRNA synthetases may have been more complex than was previously apparent (31).
Evolution of tRNA Recognition in the AspRSs Is Complex.
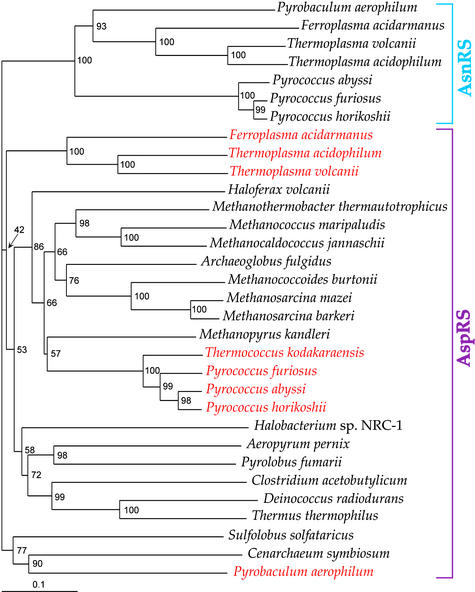
The ease of in vitro conversion of the tRNA discriminating nature of T. kodakaraensis AspRS suggests that changes between D-AspRS and ND-AspRS may have evolved more often than expected. Thus, the number of at least three independent groups of discriminating enzymes revealed by the phylogeny of the archaeal-type AspRSs (Fig. 5) may not be surprising. It has been argued that the ancestral AspRS was nondiscriminating (33), allowing for the later emergence of an AsnRS (11, 31) and a D-AspRS. However, an evolutionary change from D-AspRS to ND-AspRS may have also been logical, if one considers that Asp-tRNAAsn may not be as toxic to cellular protein synthesis as had been expected (B. Min, personal communication). The presence of a ND-AspRS may be beneficial in other ways, for instance, in case an organism needs to make asparagine (13). Therefore, it may be advantageous for an organism to retain “flexibility” with tRNA discrimination conversions in either direction, possibly in response to codon-usage drift or changing metabolic environments. Establishing a more complete picture of the dynamic evolution of Asp-tRNA and Asn-tRNA formation (31) will require further surveys of tRNA discrimination by AspRS enzymes from many more organisms.
Figure 5.
Phylogeny of archaeal-type AspRS (purple) and archaeal AsnRS (blue) proteins inferred by using the neighbor-joining method. Organisms containing AspRS proteins predicted or demonstrated to be discriminating (ref. 15) are red. Bootstrap percentages are given for each branch. Sequences were aligned with CLUSTAL X (ref. 32). (Bar, 10 amino acid replacements per 100 positions.)
Acknowledgments
We thank Alexandre Ambrogelly, Hannah Geller, Carla Polycarpo, Jesse Rinehart, Benfang Ruan, and Juan Salazar for materials and experimental help; Rick Cavicchioli, Edward DeLong, and Christa Schleper for providing AspRS sequences before publication; Karl Stetter for P. furiosus cells; and Jeffrey L. Hansen for help with structural modeling. L.F. is a National Institutes of Health postdoctoral fellow. This work was supported by grants from the National Institute of General Medical Sciences, the Department of Energy, and the National Aeronautics and Space Administration.
Abbreviations
- AspRS
aspartyl-tRNA synthetase
- D-AspRS
discriminating AspRS
- ND-AspRS
nondiscriminating AspRS
- AARS
aminoacyl-tRNA synthetase
- AsnRS
asparaginyl-tRNA synthetase
References
- 1.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 3.Becker H D, Kern D. Proc Natl Acad Sci USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curnow A W, Tumbula D L, Pelaschier J T, Min B, Söll D. Proc Natl Acad Sci USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lapointe J, Duplain L, Proulx M. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raczniak G, Becker H D, Min B, Söll D. J Biol Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 7.Curnow A, Ibba M, Söll D. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 8.Curnow A W, Hong K W, Yuan R, Kim S I, Martins O, Winkler W, Henkin T M, Söll D. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tumbula D L, Becker H D, Chang W-Z, Söll D. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 10.Thanbichler M, Böck A. EMBO J. 2002;21:6925–6934. doi: 10.1093/emboj/cdf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woese C R, Olsen G J, Ibba M, Söll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker H D, Roy H, Moulinier L, Mazauric M H, Keith G, Kern D. Biochemistry. 2000;39:3216–3230. doi: 10.1021/bi992573y. [DOI] [PubMed] [Google Scholar]
- 13.Min B, Pelaschier J T, Graham D, Tumbula-Hansen D, Söll D. Proc Natl Acad Sci USA. 2002;99:2678–2683. doi: 10.1073/pnas.012027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt E, Moulinier L, Fujiwara S, Imanaka T, Thierry J C, Moras D. EMBO J. 1998;17:5227–5237. doi: 10.1093/emboj/17.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumbula-Hansen D, Feng L, Toogood H, Stetter K O, Söll D. J Biol Chem. 2002;277:37184–37190. doi: 10.1074/jbc.M204767200. [DOI] [PubMed] [Google Scholar]
- 16.Eiler S, Dock-Bregeon A, Moulinier L, Thierry J C, Moras D. EMBO J. 1999;18:6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Briand C, Poterszman A, Eiler S, Webster G, Thierry J, Moras D. J Mol Biol. 2000;299:1051–1060. doi: 10.1006/jmbi.2000.3819. [DOI] [PubMed] [Google Scholar]
- 18.Cavarelli J, Rees B, Ruff M, Thierry J C, Moras D. Nature. 1993;362:181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- 19.Giegé R, Sissler M, Florentz C. Nucleic Acids Res. 1998;22:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pütz J, Puglisi J D, Florentz C, Giegé R. Science. 1991;252:1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- 21.Becker H D, Giegé R, Kern D. Biochemistry. 1996;35:7447–7458. doi: 10.1021/bi9601058. [DOI] [PubMed] [Google Scholar]
- 22.Nakatani M, Ezaki S, Atomi H, Imanaka T. J Bacteriol. 2000;182:6424–6433. doi: 10.1128/jb.182.22.6424-6433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varshney U, Lee C P, RajBhandary U L. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 24.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara S, Takagi M, Imanaka T. Biotechnol Annu Rev. 1998;4:259–284. doi: 10.1016/s1387-2656(08)70073-5. [DOI] [PubMed] [Google Scholar]
- 26.Eriani G, Dirheimer G, Gangloff J. Nucleic Acids Res. 1990;18:7109–7118. doi: 10.1093/nar/18.23.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christopher J A. spock, The Structural Properties Observation and Calculation Kit (Center for Macromolecular Design, Texas A&M Univ., College Station) 1998. [Google Scholar]
- 28.Liu D R, Magliery T J, Pastrnak M, Schultz P G. Proc Natl Acad Sci USA. 1997;94:10092–10097. doi: 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakasugi K, Quinn C L, Tao N, Schimmel P. EMBO J. 1998;17:297–305. doi: 10.1093/emboj/17.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekine S, Nureki O, Shimada A, Vassylyev D G, Yokoyama S. Nat Struct Biol. 2001;8:203–206. doi: 10.1038/84927. [DOI] [PubMed] [Google Scholar]
- 31.Shiba K, Motegi H, Yoshida M, Noda T. Nucleic Acids Res. 1998;26:5045–5051. doi: 10.1093/nar/26.22.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 33.Di Giulio M. J Mol Evol. 2002;55:616–622. doi: 10.1007/s00239-002-2357-6. [DOI] [PubMed] [Google Scholar]