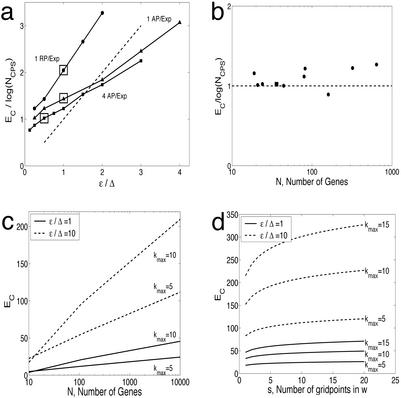
Figure 4.
Scaling behavior of the required number of perturbation experiments to identify a gene regulatory network. (a) Increasing the error ɛ increases the number of critical experiments (EC). The experimental resolution is reduced (increased ɛ) by using the same set of simulated expression data; thus, the signal (Δ) is unchanged. The estimated slopes, the required number of experiments EC scaled by log(NCPS), from Fig. 3 are plotted (boxed) for reference (RP, randomly selected perturbation; AP, algorithmically selected perturbation). For reference, a dashed line with unity slope is plotted (n = 40). (b) Scaling behavior with increasing network size (N). Multiple toggles (four to eight) are used with ɛ/Δ = 0.5. The filled box indicates the n = 40 case by using four algorithmically selected perturbations per experiment. (c) Theoretical estimates of the critical number of experiments, EC, as a function of N, maximal number of inputs kmax, and two different values of the ɛ/Δ term. (d) Small increase in EC with increasing resolution in w (i.e., s) for two different values of the ɛ/Δ term (n = 2,000).