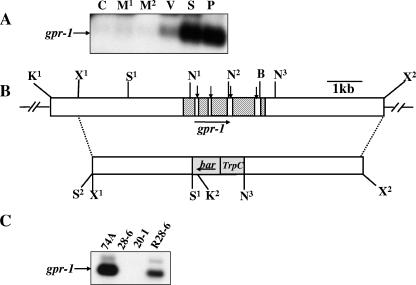
FIG. 2.
Structure of the N. crassa gpr-1 genomic region and construction of a Δgpr-1 strain. (A) Expression of gpr-1 during the N. crassa life cycle. Total RNA was isolated from cultures of wild-type strain 74A and subjected to quantitative RT-PCR analysis as described in Materials and Methods. The tissues used were the following: C, conidia; M1, 8-h submerged (mycelial) cultures; M2, 16-h submerged (mycelial) cultures; V, asexual vegetative cultures grown for 3 days at 30°C in the dark on solid VM medium; S, cultures grown for 6 days on SCM plates at 25°C under light (protoperithecial cultures); and P, SCM plate cultures 3 days after fertilization with wild-type strain 74a conidia (perithecial cultures). (B) gpr-1 genomic clone and gene replacement vector. The hatched region (top bar) indicates the gpr-1 exons, while the vertical arrows indicate introns (white areas between exons). The gray area (bottom bar) corresponds to the phosphinothricin-resistance gene, bar, under control of the A. nidulans trpC promoter. The dashed lines illustrate the homologous recombination event leading to replacement of the gpr-1 ORF with the bar cassette between the SalI (S1) and the third NcoI sites. The horizontal arrows show the direction of transcription of gpr-1 and bar. Restriction sites are as follows: N, NcoI; S, SalI; K, KpnI; B, BamHI; and X, XbaI. The second SalI site (S2) is an artifact of cloning. (C) Verification of the gpr-1 deletion mutant and complemented strains. Total RNA was isolated from 6-day-old protoperithecial tissues of wild type (74A), homokaryotic Δgpr-1 mutants (28-6 and 20-1), and a Δgpr-1 + gpr-1+ complemented strain (R28-6). Levels of gpr-1 mRNA were quantitated using RT-PCR as described in Materials and Methods. The faint, larger molecular mass species in the 74A and R28-6 samples is due to amplification of residual genomic DNA in the RNA preparations.