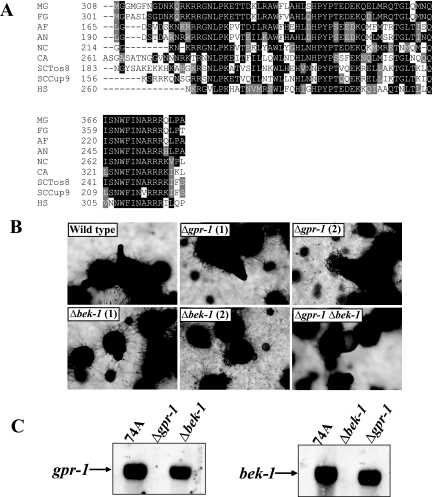
FIG. 6.
Epistasis analysis between gpr-1 and bek-1. (A) Alignment of BEK-1 with related homeodomain transcription factors. ClustalW (http://www.ch.embnet.org/software/ClustalW.html) was used to align N. crassa BEK-1 (NC; NCU00097.2) with putative homeodomain transcription factors from the fungi F. graminearum (FG; FG05475.1), A. fumigatus (AF; Afu4g10110), A. nidulans (AN; AN2020.2), and C. albicans (CA; CaJ7.0235) and previously characterized homeodomain proteins identified as M. grisea Pth12p (MG; DQ060925.1), S. cerevisiae Tos8p (SCTos8; YGL096W) and Cup9p (SCCup9; YPL177C), and human PKNOX1 (HS; accession number AAC51243). Boxshade was used to indicate identical (black shading) and similar (gray shading) amino acid residues (http://www.ch.embnet.org/software/BOX_form.html). (B) Perithecial development. Perithecia from wild-type (74A), Δgpr-1 (28-6), Δbek-1 (BK1), and Δgpr-1 Δbek-1 (GP1BK1) strains were analyzed microscopically 6 days after fertilization. Images were captured at ×52 magnification. Two images were taken for Δgpr-1 and Δbek-1 strains, showing perithecial defects (1) and ruptured perithecia (2). (C) Expression of gpr-1 and bek-1. Total RNA was isolated from perithecial tissues of wild-type (74A), Δgpr-1 (28-6), and Δbek-1 (BK1) strains 3 days after fertilization and subjected to quantitative RT-PCR as described in Materials and Methods.