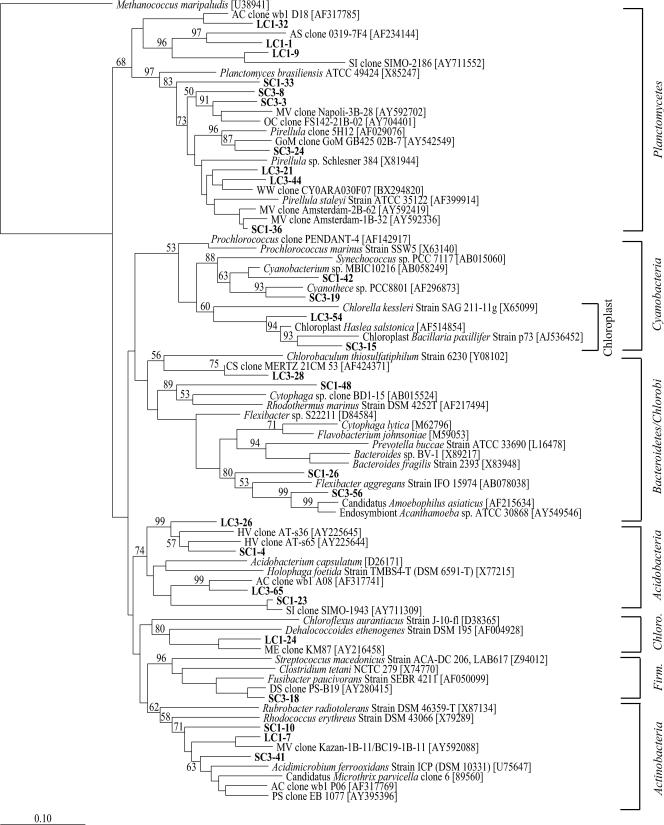
FIG. 4.
Nonproteobacterial neighbor-joining phylogenetic tree, incorporating a Jukes-Cantor distance correction, of SSU rRNA genes from SC#1, SC#3, LC#1, LC#3, and SAB samples. Sequences from this study and close relatives were aligned using the Fast Aligner algorithm, verified by hand, and compared to the E. coli SSU rRNA secondary structure using the ARB software package. Bootstrap analyses were conducted on 1,000 samples, and percentages greater than 50% are indicated at the nodes. Methanococcus maripaludis was used as the outgroup. Bar, 0.10 change per nucleotide position.