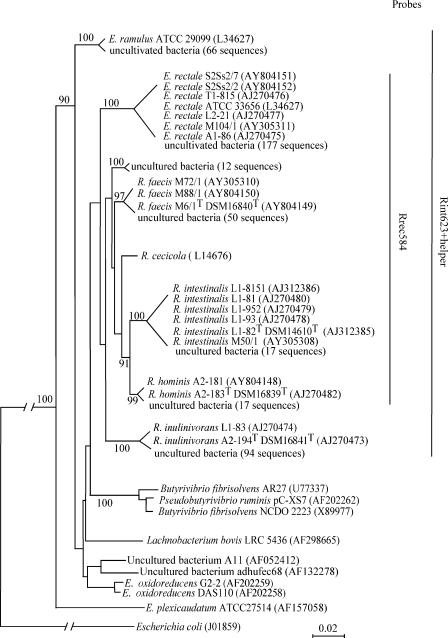
FIG. 1.
Phylogenetic tree, constructed by the neighbor-joining method, based on 16S rRNA sequences of Roseburia- and E. rectale-related clones and species. Isolation of strains T1-815, L2-21, A1-86, L1-8151, L1-81, L1-82T, L1-952, L1-93, A2-181, A2-183T, L1-82, and A2-194T is described in reference 23, and isolation of strains M104/1, M72/1, M88/1, M6/1T, and M50/1 is described in reference 23; for S2Ss2/7 and S2Ss2/2, see the text. 16S rRNA sequence accession numbers are given in parentheses. Numbers above each node are confidence levels (percent) generated from 1,000 bootstrap trials. The Escherichia coli sequence is used as the outgroup to root the tree. The scale bar refers to fixed nucleotide substitutions per sequence position. For the sake of space, the tree is presented in a schematic form that retains the general topology. The branch length of E. coli is not to scale. The probe coverage is shown on the extreme right.