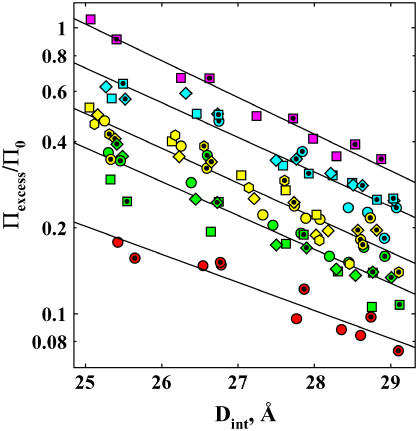
FIGURE 3.
Dependence of the change in excess water for Spd3+-DNA, Πexcess/Π0, on interhelical spacing is shown for a set of 12 nonpolar alcohols. Condensed DNA arrays were equilibrated against 2 mM SpdCl3 and 10 mM TrisCl (pH 7.5), PEG, and varied concentrations of alcohols, at 20°C. The alcohols are coded such that solutes with the same excess number of alkyl carbons over hydroxyl groups have the same color. The alcohols are methanol (1.5 and 3 molal), red circles; ethanol (1 and 2 molal), green circles; 1,3-propanediol (1 and 2 molal), green squares; 1,2-propanediol (1 and 2 molal), green diamonds; 1-propanol (2 molal), yellow circles; 2-propanol (1 and 2 molal); yellow squares; 1,4-butanediol (1 and 2 molal), yellow diamond; 2,3-butanediol (1 and 2 molal), yellow hexagon; 1-butanol (0.5 and 1 molal), aqua circles; 2-butanol (0.5 and 1 molal), aqua squares; t-butanol (0.5 and 1 molal)-(0.5 and 1 molal), aqua diamonds; and MPD (0.5 and 1 molal), pink square. Data for the lower alcohol concentration are given by the symbols with inner dots. The decay length of the apparent exponential, calculated from linear fits on the logarithmic scale (solid lines), varies between ∼3.5 and 4 Å. The amplitude or preexponential factor varies significantly with the chemical nature of the osmolyte, but not its size.