Abstract
Viral inclusion bodies, or viroplasms, that form in rotavirus-infected cells direct replication and packaging of the segmented double-stranded RNA (dsRNA) genome. NSP2, one of two rotavirus proteins needed for viroplasm assembly, possesses NTPase, RNA-binding, and helix-unwinding activities. NSP2 of the rotavirus group causing endemic infantile diarrhea (group A) was shown to self-assemble into large doughnut-shaped octamers with circumferential grooves and deep clefts containing nucleotide-binding histidine triad (HIT)-like motifs. Here, we demonstrate that NSP2 of group C rotavirus, a group that fails to reassort with group A viruses, retains the unique architecture of the group A octamer but differs in surface charge distribution. By using an NSP2-dependent complementation system, we show that the HIT-dependent NTPase activity of NSP2 is necessary for dsRNA synthesis, but not for viroplasm formation. The complementation system also showed that despite the retention of the octamer structure and the HIT-like fold, group C NSP2 failed to rescue replication and viroplasm formation in NSP2-deficient cells infected with group A rotavirus. The distinct differences in the surface charges on the Bristol and SA11 NSP2 octamers suggest that charge complementarity of the viroplasm-forming proteins guides the specificity of viroplasm formation and, possibly, reassortment restriction between rotavirus groups.
A characteristic of many viral infections is the formation of large inclusion bodies, or viroplasms, within the cell that serve as sites of genome replication and capsid morphogenesis. Although the complex protein-protein and protein-RNA interactions that make up these viral factories are poorly defined, they must provide a microenvironment with the necessary fluidity required for replication and particle-assembly processes to proceed efficiently while supporting the structural integrity of the inclusion. In the case of rotaviruses, members of the Reoviridae, two nonstructural proteins, NSP2 and NSP5, interact to form a membrane-free viroplasm scaffold (12) in which the segmented double-stranded RNA (dsRNA) of the virus is replicated and packaged into previrion core particles (32). X-ray crystallographic studies have shown that NSP2 self-assembles into large doughnut-shaped octamers, through tail-to-tail stacking of tetramers (18). Besides having affinity for the dimeric phosphoprotein NSP5 (10), the NSP2 octamer also possesses nonspecific RNA-binding activity (34), possibly mediated by the highly electropositive grooves that extend diagonally across the tetramer-tetramer interface of the octamer. NSP2 possesses an Mg2+-dependent hydrolysis activity, capable of removing the γ-phosphate from nucleoside triphosphates (NTPs) and thus serving as an NTPase (34). This hydrolytic activity is mediated by residues in the cleft region of each NSP2 monomer that form a histidine triad (HIT)-like motif (4), an element common to a large and ubiquitous family of nucleotidyl hydrolases (2) that heretofore has not been seen in a viral protein. Although the unique structural and enzymatic properties of the octamer can be anticipated to be important for virus replication, how these features contribute to viroplasm formation and to processes occurring within the viroplasm has not been resolved.
The rotavirion is an icosahedron consisting of three concentric protein layers that encapsidates 11 segments of dsRNA. The antigenicity of the protein that forms the intermediate layer (VP6) is used to classify rotaviruses into groups (e.g., A, B, and C) (11). Virus strains belonging to the same group can undergo genetic recombination (reassortment) upon coinfection, while strains belonging to different groups cannot. The basis of reassortment restriction between groups is undefined. During cell entry, rotavirions are converted to transcriptionally active double-layered particles that synthesize the template positive-strand RNAs for translation and genome replication. The positive-strand RNAs are packaged into and replicated by core replication intermediates in viroplasms (28). These cores represent icosahedral T = 1 particles surrounded by the core lattice protein, VP2, that contain minor amounts of the viral RNA-dependent RNA polymerase (RdRP), VP1, and the 5′-capping enzyme, VP3. NSP2 is associated with these intermediates, possibly through its affinity for single-stranded RNA (ssRNA) or the RdRP.
Although much has been learned about NSP2 through studies on the purified recombinant form of the protein, our understanding of the role of NSP2 in the biological system is limited. Some insight has been gained by analysis of cells infected with the tsE(1400) strain, a mutant SA11 rotavirus with a temperature-sensitive (ts) lesion mapping to the genome segment (gene 8) encoding NSP2 (29). Notably, at the nonpermissive temperature, tsE-infected cells are defective in viroplasm formation, synthesize low levels of viral dsRNA, and overaccumulate empty particles (6). RNA interference experiments have shown similar changes in infected cells in which NSP2 expression has been suppressed (32). These findings suggest a role for NSP2 not only in viroplasm formation, but also in coordinating genome packaging and replication with particle assembly. The critical role of NSP2 in viroplasm formation has been confirmed by transient expression studies, which have shown that the expression of NSP2 in combination with NSP5 promotes assembly of viroplasm-like structures (VLS) (12).
To further understand the role of NSP2 in rotavirus replication, we have initiated a series of structural and biochemical studies designed (i) to address whether the architecture of NSP2 monomer and octamer is conserved across the various groups of rotavirus; (ii) to identify properties of heterologous NSP2 and NSP5 required in support of genome replication and viroplasm formation; and (iii) to define how the various functional properties of NSP2, such as NTPase and RNA-binding activities and viroplasm formation, are partitioned structurally. In this study, we have determined the atomic structure of NSP2 encoded by the group C Bristol strain of rotavirus (NSP2c), which exhibits only 35% sequence identity with group A NSP2, and compared it to that encoded by the group A SA11 strain (NSP2a). Although the results revealed extensive conservation of octamer architecture, significant differences were noted in the overall surface potential of the octamers. To explore the contribution of the conserved structural elements to the function of NSP2 in the context of infection, we created a novel NSP2-dependent complementation system that supports genome replication and particle assembly in tsE-infected cells containing a gene 8-specific siRNA, but only when NSP2a is expressed in trans from siRNA-resistant transcripts. Using this assay system, we demonstrate that the HIT-dependent NTPase activity of NSP2a is necessary for dsRNA synthesis but not for viroplasm formation and that despite overall structural homology, NSP2c fails to functionally substitute for NSP2a in supporting viral replication and this failure is likely to due to significant differences in the surface charge distribution.
MATERIALS AND METHODS
Expression and purification of NSP2.
The NSP2 cDNA in the plasmid RotaC seg9/M13 of the Bristol strain (7) was amplified by PCR and cloned into the bacterial expression vector pQE60 (pQE60g8C), such that six His residues were fused to the 3′ end of the NSP2c open reading frame (ORF). Bacterially expressed NSP2c was initially purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography as described for SA11 NSP2 (NSP2a) (34). Polyclonal antisera to NSP2c were produced in guinea pigs by immunization with 50 μg of protein in Freund's complete adjuvant and again in incomplete adjuvant at weeks 2, 4, and 6.
Preparation of NSP2 for crystallization.
NSP2a was expressed in Escherichia coli strain SG13009 (QIAGEN) and purified as described before (18). NSP2c was expressed and purified in the same manner with slight modification. E. coli SG13009 cells containing pQE60g8C were grown to an optical density of 0.7 at 600 nm in Terrific Broth (Invitrogen), when the expression of NSP2c was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After incubation for 8 to 10 h at 37°C, the bacteria were recovered by centrifugation at 4,200 × g for 30 min. The cell pellet from a 4-liter culture was resuspended in 200 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0, 40 μg/ml RNase A) and processed twice using a Microfluidizer high-shear processor (model 110Y; Microfluidics, Newton, Mass.). To recover His-tagged NSP2c, the clarified lysate was incubated with 8 ml of Ni-NTA agarose beads (QIAGEN). Subsequently, the beads were washed four times with buffer containing 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0, and once with the same buffer, except containing 50 mM imidazole. Protein was eluted with 70 ml of the same buffer containing 250 mM imidazole, and then using a Centricon centrifugation cartridge, the buffer was changed to 10 mM Tris-100 mM NaCl, pH 8.0, with a protein concentration of 4 to 6 mg/ml.
Crystallization.
NSP2a was crystallized as described before (18). NSP2c was crystallized by hanging drop vapor diffusion. Microseeding technique was used to grow crystals of better quality. Drops were assembled with 4 μl of protein mixed with 4 μl of well solution [1.5 to 1.7 M (NH4)2SO4, 100 mM morpholineethanesulfonic acid (MES), pH 6.4], and then preequilibrated overnight. A very small amount of solution from a drop in which a crystal had been crushed was placed at one point of the overnight-preequilibrated protein drop mixture without mixing. Crystals typically grew after several hours.
Data collection and processing.
NSP2c crystals were suspended in mother liquor with 0.25 M and then 0.5 M Li2SO4 for 1 min each and transferred into 1 M Li2SO4. Crystals were then plunge-frozen in liquid nitrogen. Diffraction data were collected on SBC-CAT beam line 19ID at the Advanced Photon Source (Argonne National Laboratory). Data were processed using the HKL2000 (26) software package. Diffraction data for NSP2a crystals under cryo-conditions were collected on the F2 beam line at CHESS (Cornell University).
Structure determination and refinement.
The structures of NSP2c and NSP2a (to 2.2-Å resolution) were solved by molecular replacement methods using the previously published 2.6-Å structure of NSP2a as a search model. The molecular replacement solution was obtained using PHASER (24). Model building was carried out manually using the program O (19). Any model bias was examined by computing composite-OMIT maps using CNS (3). Iterative rounds of refinement with simulated annealing in CNS and model building with O were carried to obtain the final model. Details of the diffraction data analysis and structure refinement are provided in Table S1 of the supplemental material. Optimized multiple superposition was obtained with LSQMAN in the CCP4 suite of programs (9). An electrostatic potential map of the octamer was calculated using Delphi (17). Figures were generated with PYMOL (http://www.pymol.org). The accession code for NSP2c in the Protein Data Base is 2GUO.
Expression vectors.
Wild-type (wt) and mutated NSP2a ORFs were prepared by PCR and subcloned in the T7 transcription vector, SP72. The g8* nomenclature identifies pSP72 with NSP2a-encoding gene 8 cDNAs that have been mutated in the target site for the g8D siRNA, such that the siRNA no longer induces degradation of transcripts made from the vector. The mutations were introduced into the codon wobble positions of the target site and did not alter the NSP2a amino acid sequence. The g8D siRNA target site in gene 8 cDNAs was mutated with a QuikChange kit (Stratagene) and the overlapping primer pair 5′-GATGTTCATGTTAAGGAGCTAGTGGCAGAGCTGCGATGGCAATATAAC-3′ and 5′-GTTATATTGCCATCGCAGCTCTGCCACTAG CTCCTTAACATGAACATC-3′.
To prepare the T7 transcription vectors pSP72g8*/D36A and pSP72g9C, the NSP2 coding region in pQE60g8/D36A and pQE60g9C was amplified using an Expand high-fidelity PCR kit. The amplified products were digested with EcoRI and XhoI and inserted into the corresponding restriction sites of pSP72. The vectors pSP72g8*-HA and pSP72g8*/K188A-HA were generated by outward PCR using an Expand high-fidelity PCR kit. The amplified DNAs were digested with HindIII and then self-ligated to produce plasmids. The vectors pSP72g8*ΔC4 and pSP72g8*ΔC7 were similarly generated using PCR. The amplified DNAs were digested with EcoRI and XhoI and ligated into corresponding sites of pSP72.
siRNAs and transfection of cells.
The g8D and irrelevant (IR) siRNAs (Dharmacon Research) were described previously (32). The control IR siRNA targets the Wa gene 5 RNA, but not any of the SA11 RNAs.
Fetal rhesus monkey kidney (MA104) cells grown in 12-well plates were rinsed with minimal essential medium (MEM) and then overlaid with 1 ml of (Quality Biological Inc., Gaithersburg, MD) MEM per well. Transfections were carried out by adding to each well a 200-μl volume of Opti-MEM I containing 2% Lipofectamine 2000 (Invitrogen), 0.5 μg of the appropriate T7 transcriptional vector, and/or 0.3 μM siRNA. After incubation for 4 h, 1 ml of MEM containing 20% fetal bovine serum was added to each well. Incubation was then continued for an additional 2 h, at which point cells were infected.
Virus infection.
Transfected cells were infected with trypsin-activated tsE strain and/or vaccinia virus vTF7.3 (13), each at a multiplicity of infection (MOI) of 10. At 1 h postinfection (p.i.), the inoculum was replaced with MEM containing 40 μg of cytosine arabinoside and 100 μg of rifampin (Sigma) per ml. To radiolabel proteins, the inoculum was replaced instead with 80% methionine- and cysteine-free MEM and 20% MEM, containing 25 μCi of [35S]-Express protein labeling mix (Perkin-Elmer Life Sciences) per ml. To radiolabel RNAs, the inoculum was replaced with 80% phosphate-free Dulbecco's MEM and 20% MEM supplemented with 25 μCi of 32Pi (8,000 to 9,000 Ci/mmol) per ml. Cells were typically harvested at 12 h p.i.
Protein analysis.
Infected cell lysates were analyzed by electrophoresis on 10% NuPAGE Bis-Tris gels (Invitrogen). Proteins were detected by autoradiography. In Western blot assays, proteins were detected on nitrocellulose membranes by incubation with a 1:500 dilution of guinea pig antisera prepared against NSP2a or NSP2c. Hemagglutinin (HA)-tagged protein was detected with rat anti-HA monoclonal antibody (1:2,000). Horseradish peroxidase-conjugated goat anti-guinea pig, anti-mouse, and anti-rat antisera were used as secondary antibodies (1:10,000). Blots were developed with SuperSignal West Pico chemiluminescent substrate (Pierce).
RNA analysis.
Infected cells were lysed by freeze-thawing in water and centrifuged to remove large cellular debris. After treatment with 160 U of DNase I (Invitrogen) per ml for 30 min at 37°C, RNA was purified by phenol-chloroform extraction, resolved by electrophoresis on 12% polyacrylamide gels, detected by autoradiography, and quantified using a phosphorimager. The levels of dsRNA synthesis were compared among samples by measuring the intensity of gene 1 after correcting for background in the corresponding lanes.
Isolation of virus particles.
Infected MA104 cells labeled with 35S-labeled amino acids were scraped into the media at 24 h p.i. and lysed by freeze-thawing. After removal of cell debris by low-speed centrifugation, virus was pelleted by centrifugation at 160,000 × g for 2 h. The pellet was suspended in Tris-buffered saline (TBS) buffer (25 mM Tris-HCl, pH 7.4, 137 mM NaCl, 5 mM KCl, 1 mM MgCl2, 0.7 mM CaCl2, 0.7 mM Na2HPO4, 5.5 mM dextrose), and virus was banded by CsCl centrifugation. Fractions (0.5 ml) from the gradient were analyzed by electrophoresis on 10% NuPAGE Bis-Tris gels and autoradiography.
IFF assay.
The infectious focus-forming (IFF) assay was performed as follows. CsCl gradient fractions containing triple-layered particles were pooled and treated with 5 μg of trypsin per ml. The samples were diluted with medium 199 (Invitrogen), and virus in the dilutions was adsorbed onto monolayers of MA104 cells. The inoculum was replaced with medium 199 containing 40 μg of cytosine arabinoside, 100 μg of rifampin (Sigma), and 10 μg of actinomycin D (Invitrogen) per ml to inhibit the growth of vaccinia virus. The cells were maintained at 31°C for 30 h, washed three times with phosphate-buffered saline (PBS), and fixed by incubation for 2 min in methanol. The cells were immunostained by sequential incubation with guinea pig anti-VP6 polyclonal sera (1:2,000) and horseradish peroxidase-conjugated goat anti-guinea pig antibody (1:3,000), each for 30 min at room temperature. The cells were washed four times with PBS prior to developing for 20 min in a solution containing 0.266 mg per ml of 3-amino-9-ethylcarbazole in 0.1 M sodium acetate, pH 5.2, and 0.045% hydrogen peroxide. Stained cells were counted with an inverted light microscope.
IF analysis.
MA104 cells were infected at an MOI of 10 with vTF7.3 and/or the tsE strain. At 1 h p.i., the inoculum was removed and replaced with a mixture of 4% Lipofectamine (Invitrogen) and 0.5 μg each of one or more T7 transcription vectors in medium 199. At 6 h p.i., the transfection mixture was replaced with medium 199 containing 10% FBS. The cells were processed for immunofluorescence (IF) at 16 h p.i. by fixing with formaldehyde and permeabilizing with Triton X-100 (4). Afterwards, the cells were incubated with a 1:500 to 1:1,000 dilution of one or more of the following: guinea pig polyclonal antisera prepared against recombinant NSP2a or NSP2c, rat anti-HA monoclonal antibody, and mouse anti-NSP5 monoclonal antibody 158G37. After washing, the cells were incubated with a 1:1,000 dilution of one or more of the following: Alexa Fluor 488 goat anti-guinea pig immunoglobulin G (IgG), Alexa Fluor 594 goat anti-mouse IgG, and Alexa Fluor 568 goat anti-rat IgG (Molecular Probes). Nuclei were detected by staining with Hoechst 33258 (4′,6′-diamino-2-phenylindole [DAPI]).
Protein structure accession number.
The atomic coordinates for NSP2c have been deposited in the Protein Data Bank (PDB) under accession code 2GUO.
RESULTS
Structure of NSP2c.
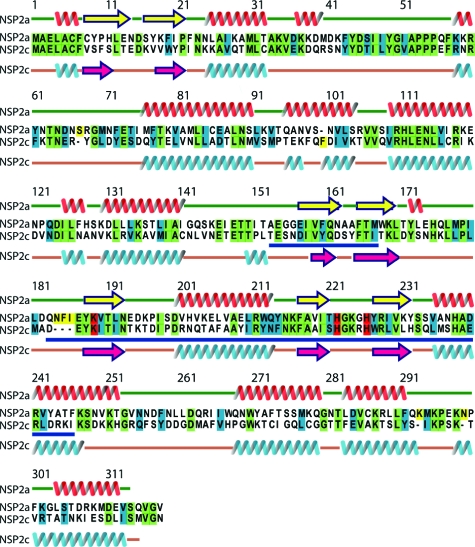
The group A rotaviruses, which include the prototypic strain SA11, are a widespread cause of life-threatening dehydrating diarrhea in young children (21). In contrast, group C rotaviruses are generally associated with limited outbreaks of diarrheal disease. The group C Bristol strain originated from a family outbreak of gastroenteritis that resulted in the death of a young child (5). Sequencing shows that NSP2c shares limited similarity (57%) and identity (35%) with NSP2a (Fig. 1). As an approach for identifying structural features of NSP2 that are vital to rotavirus replication, we determined the atomic structure of NSP2c and contrasted its architectural features with those of NSP2a.
FIG. 1.
Sequence alignment of NSP2a with NSP2c. Identical, similar, and catalytic residues are highlighted in green, cyan, and red, respectively. The secondary structural elements as observed in the X-ray structures of NSP2a and NSP2c are indicated above and below their respective sequences. The HIT homology region is underlined in blue. Sequence alignment was carried out using the program ClustalW. Sequences shown are from the SwissProt Database: NSP2a, accession no. Q03243; and NSP2c, accession no. Q9PY93.
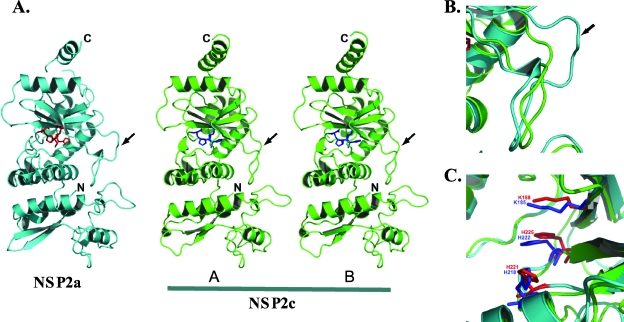
Recombinant NSP2c with a C-terminal His tag was expressed in bacteria and purified under conditions similar to NSP2a (18). NSP2c crystallized in space group I4 with two molecules in the asymmetric unit of the crystal, in contrast to NSP2a (18), which crystallized in space group I422 with one molecule in the asymmetric unit. Using the X-ray structure of NSP2a (PDB identification no. 1L9V) as an initial phasing model for molecular replacement, we determined the X-ray structure of NSP2c to a 2.8-Å resolution (Fig. 2). After several iterative cycles of model building and refinement, the structure was refined with a final R factor of 0.23 (Rfree = 0.281). Residues Ala 2 to Lys 246 and Tyr 254 to Met 309 from each molecule could be built into the electron density. Recently, we obtained crystals of NSP2a that diffracted to a 2.2-Å resolution (Fig. 2). The structure of NSP2a was determined to 2.2 Å using the previously published 2.6-Å structure (PDB identification no. 1L9V) with an overall R factor of 0.233 (Rfree = 0.268). The refined 2.2-Å structure consists of residues Ala 2 to Pro 295 and Pro 299 to Val 312. The 2.2-Å NSP2a structure is essentially the same as the 2.6-Å structure, except that some loops are better defined and several water molecules are easily identified.
FIG. 2.
Comparison of the monomeric subunits of NSP2a and NSP2c. (A) Ribbon diagram of NSP2a (cyan) and the two molecules (A and B) in the crystallographic asymmetric unit of NSP2c (green). The locations of the N and C termini are denoted. The catalytic residues of NSP2a (His 225, His 221, and Lys 188) are shown in red, and those of NSP2c (His 222, His 218, and Lys 185) are shown in blue. The loop (residues 170 to 185) is shown by an arrow. (B) A close-up view of the superposition of the loop regions showing noticeable differences between NSP2a and NSP2c structures. In both panels B and C, the same color scheme as in panel A is used. (C) A close-up view of the superposition of catalytic residues in NSP2a and NSP2c structures.
Despite only 35% identity, the overall tertiary structure of the NSP2c monomer is very similar to that of the NSP2a monomer with distinct N-terminal and C-terminal domains separated by a deep cleft (Fig. 1 and 2). The root mean square deviation between the two structures is 1.8 Å for the matching Cα atoms. Located in the cleft are the residues forming a HIT-like motif implicated in nucleotide binding and hydrolysis (4). When the structures of the NSP2a and NSP2c monomers are superimposed, the proposed catalytic residues of the cleft (His 225 and His 221 of NSP2a and His 222 and His 218 of NSP2c) are closely matched (Fig. 2C), indicating a conservation of the active site.
The major difference between the NSP2a and NSP2c monomers concerns the loop formed by residues 170 to 185 (Fig. 2B). In particular, NSP2c lacks residues corresponding to residues 183 to 185 of NSP2a, thus leaving NSP2c with a shorter loop structure (arrow in Fig. 2A and B). This difference appears to influence the overall positioning of the C-terminal domain, with noticeable displacements in the positions of four α-helices in the C-terminal domain of NSP2c. In the NSP2c crystal, the two molecules (A and B) in the asymmetric unit have nearly identical structures, superimposing with a root mean square deviation of 0.58 Å for matching Cα atoms. The main difference, however, between the two monomeric forms in the NSP2c asymmetric unit is that, in molecule A, the C-terminal domain is moved slightly further away from the N-terminal domain. This domain movement appears to be facilitated by the small changes within the same loop region (residues 170 to 182) that distinguish the NSP2a and NSP2c monomers.
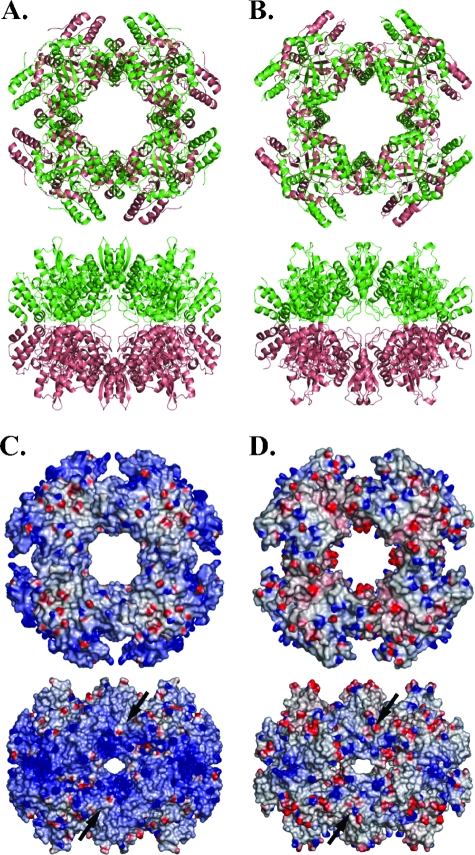
Despite different space groups, both NSP2a and NSP2c form octameric structures with tail-to-tail stacking of two four-fold related tetramers using crystallographic symmetry (Fig. 3A and B). For NSP2a, the octamer represents a 422 (d4) crystallographic symmetry, whereas for NSP2c the octamer represents only four-fold (c4) symmetry. The two octamers are essentially of the same dimensions with NSP2c octamer being slightly shorter by about ∼5 Å along the four-fold axis, and wider by the same amount along the axis perpendicular to the four-fold axis. Comparison of the NSP2 octamers by superposition of the Cα backbone of each subunit shows that their minor differences reflect the small variations described above in their monomeric subunit structures. The most striking difference between NSP2a and NSP2c octamers is in the distribution of surface electrostatic charges (Fig. 3C and D). The surface of the NSP2a octamer is significantly more basic than that of the NSP2c octamer, especially along the four grooves that run diagonally across the tetramer-tetramer interface. Based on their basic nature, the grooves were proposed to serve as the RNA-binding sites of the octamer (18). More recently, cryo-electron microscopy studies have shown that grooves not only represent RNA-binding sites but also NSP5-binding sites as well (Jiang et al., unpublished data). Notably, while each subunit of the NSP2 octamer contains an NTP-binding site, the RNA- and NSP5-binding grooves are formed by intersubunit interactions in the octamer. Thus, the ability of NSP2 to self-assemble into an octamer is critical for its interaction with RNA and NSP5.
FIG. 3.
Comparison of NSP2a and NSP2c octamers. (A) NSP2a octamer (in ribbon representation) as viewed along the four-fold axis (top) and one of the two two-fold axes (bottom) of the 422 symmetric structure. (B) NSP2c octamer in the same orientations (top and bottom). In panel B, the molecules A and B, which represent the crystallographic asymmetric unit, are shown in green and brown, respectively. In panel A, although shown in different colors, the molecules are same because of 422 symmetry. (C) Electrostatic surface potentials of NSP2a octamers in the same orientations as in panel A. (D) Electrostatic potential surfaces of NSP2c octamers in the same orientations as in panel B. Blue and red represent positive and negatively charged residues, respectively. The groove across the two-fold axis is shown by black arrows.
Novel NSP2-dependent complementation system.
Biochemical analyses have indicated that NSP2c, like NSP2a, has ssRNA-binding and NTPase activity, albeit with somewhat varying specific activities (data not shown). The conserved nature of the architecture of the NSP2a and NSP2c octamers and their NTPase and RNA-binding activities indicate that these structural and biochemical features are critical to rotavirus replication. Moreover, the extensive similarity in the features of NSP2a and NSP2c raises the possibility that one can substitute for the other during virus replication.
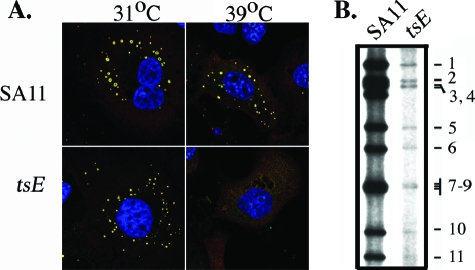
To establish a biologically relevant system to study the features of NSP2, particularly in the context of genome replication and viroplasm formation, we developed a novel complementation system wherein rotavirus replication was made dependent on the availability of NSP2 provided in trans. Two critical aspects of the system are as follows. (i) Instead of infecting cells with wt SA11, cells were infected with the tsE(1400) strain, a mutant virus encoding NSP2 with a ts mutation (A152V). At permissive temperature (31°C), the tsE strain grows to a titer that is ∼4 logs higher than at nonpermissive temperature (39°C). As shown by IF staining (Fig. 4A), tsE-infected cells maintained at 39°C are defective in viroplasm formation. Although well-developed viroplasms can be detected in tsE-infected cells maintained at 31°C, such viroplasms can be detected in SA11-infected cells maintained at either 31 or 39°C (Fig. 4A). Based on 32Pi labeling, the amount of dsRNA produced in tsE-infected cells at 39°C is ∼100-fold less than that produced at 31°C (Fig. 4B). (ii) An SA11 gene 8-specific (g8D) siRNA was transfected into tsE-infected cells to suppress the expression of virus-encoded NSP2a. In previous studies, g8D siRNA was shown to reduce NSP2a expression in SA11-infected cells by ∼10-fold, thereby inhibiting viroplasm formation (32). Thus, in this NSP2-dependent complementation system, the supply of functional NSP2a required to support dsRNA synthesis and viroplasm formation was suppressed by not only using a ts form of NSP2a, but also by using an siRNA to reduce the expression of ts NSP2a. Because the transfection efficiency of MA104 cells was limited to 80 to 90% (32), 10 to 20% of the cells in the complementation system were infected with the tsE strain, but failed to receive g8D siRNA. The presence of the tsE strain in the g8D siRNA-free cells provided the advantage of preventing the high levels of background genome replication and particle assembly that would have occurred if wt SA11 virus had been used in the complementation system instead of the tsE strain.
FIG. 4.
Viroplasm formation and dsRNA synthesis in SA11- and tsE strain-infected cells. MA104 cells infected with SA11 or the tsE strain were maintained at either 31°C or 39°C until 12 h p.i. in medium containing no label or 32Pi and processed for either IF assay or dsRNA synthesis. (A) Viroplasm formation was visualized by IF assay using primary antibodies for the detection of NSP2 (green) and NSP5 (red). Colocalization of signals is indicated by yellow, and cell nuclei were visualized by DAPI. (B) 32P-labeled viral dsRNA samples (g1 to g11) harvested from infected cells maintained at 39°C were analyzed by electrophoresis on 12% polyacrylamide gels and autoradiography.
To provide a source of recombinant NSP2a in the complementation system, we used vaccinia virus vTF7.3 (13) to synthesize NSP2a mRNAs from T7 transcription vectors transfected into tsE-infected cells. This system was selected over cytomegalovirus-driven plasmids to express proteins in vivo because it provided for more rapid and higher levels of protein expression, mimicking more closely the NSP2 expression pattern during rotavirus infection. Earlier studies have also shown that productive rotavirus replication occurs despite coinfection of cells with vaccinia virus (8, 23). Vector-derived NSP2 transcripts, unlike the parallel NSP2 transcripts generated by the virus, were not degraded by g8D siRNA-induced RISC complexes due to the introduction of silent mutations into the siRNA target sequence of the NSP2a cDNA. In this article, NSP2a originating from vector-derived transcripts resistant to degradation by g8D siRNA-induced RISC complexes is identified with an asterisk (e.g., NSP2a*).
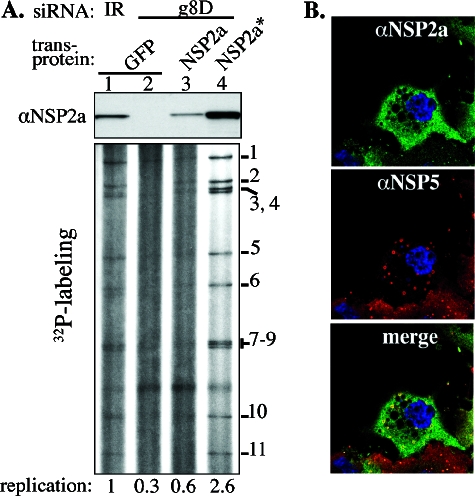
Complementation assays were performed via a two-step procedure in which MA104 cells were initially cotransfected with either g8D siRNA or an IR control siRNA and with a transcription vector specifying either NSP2 transcripts or control green fluorescent protein (GFP) transcripts. Six hours later, the cells were coinfected with the tsE strain and vTF7.3 and maintained at 39°C, sometimes in the presence of 32Pi or 35S-labeled amino acids, until 12 or more h p.i., when harvested. A comparison of the effects of transfection of various combinations of siRNAs and transcription vectors into tsE/vTF7.3-infected MA104 cells is shown in Fig. 5. As shown by Western blot assay, the g8D siRNA was efficient in knocking down the expression of NSP2a in tsE-infected cells which lacked an NSP2a transcription vector (Fig. 5A, lane 2). The loss of NSP2a was correlated with the nearly complete loss of dsRNA synthesis. Cotransfection of tsE/vTF7.3-infected cells with g8D siRNA and a vector producing g8D siRNA-sensitive transcripts resulted in low levels of NSP2a expression and little increase in levels of dsRNA synthesis (Fig. 5A, lane 3). In contrast, cotransfection with g8D siRNA and a vector producing g8D siRNA-resistant transcripts resulted in a marked increase in NSP2a* expression (Fig. 5A, lane 4) and led to levels of dsRNA synthesis exceeding those occurring in tsE/vTF7.3-infected cells containing the g8D siRNA by ∼5- to 15-fold and in cells containing IR siRNA by ∼2- to 3-fold (Fig. 5A, lane 1). Therefore, NSP2a* provided in trans compensated for the replication-deficient phenotype of tsE-infected cells in which the expression of virus-encoded NSP2a was suppressed with the g8D siRNA.
FIG. 5.
Rescue of dsRNA synthesis by NSP2*. MA104 cells were transfected with either IR or g8D siRNA; a vector expressing GFP, NSP2, or NSP2*; and infected with vTF7.3 and the tsE strain. At 12 h p.i., the infected cells were analyzed for protein expression, dsRNA synthesis, and viroplasm formation. (A) NSP2 expression was monitored by Western blot assay, and 32P-labeled viral dsRNAs from cell lysates were analyzed by electrophoresis on 12% polyacrylamide gels and autoradiography. Relative amounts of 32P-labeled dsRNA, adjusted for background, were determined using a phosphorimager. The level of replication was normalized to that in the control cells receiving IR siRNA (lane 1) and is reported as a mean of four independent quantitations with standard errors of 0.1 for the values in lanes 2 and 3 and 0.5 for the value in lane 4. αNSP2a, anti-NSP2a. (B) Viroplasms were detected in tsE strain-infected cells transfected with g8D siRNA and a T7 vector expressing NSP2* by IF assay (NSP2, green; NSP5, red; colocalization, yellow; nuclei, blue).
IF analysis showed that tsE/vTF7.3-infected cells transfected with g8D siRNA and expressing NSP2a* contained viroplasms (Fig. 5B) like those formed in SA11-infected cells (Fig. 4A). Hence, NSP2a* expression not only restored dsRNA synthesis but also promoted development of viroplasms. However, in these cells, only a fraction of the transiently expressed NSP2a* localized to viroplasms, with the majority diffusely distributed in the cytoplasm. Since NSP2a and NSP5a (i.e., NSP5 encoded by group A virus) codirect viroplasm formation, the failure of a large fraction of transiently expressed NSP2a* to localize to viroplasms may be due to the disproportionate overexpression of the plasmid-derived NSP2a* relative to virus-encoded NSP5a.
Virus assembly in NSP2*-containing cells.
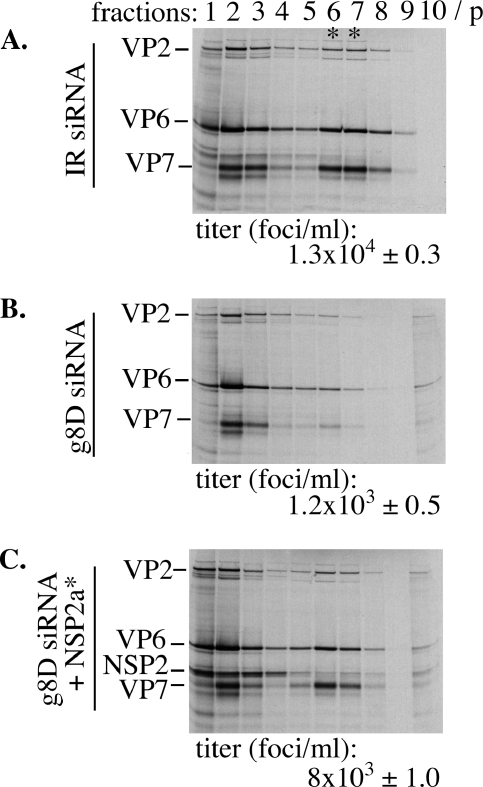
The standard method of plaque titration could not be used to measure virion assembly in complementation assays due to the presence of both the tsE strain and vTF7.3 in cell lysates. As an alternative, triple-layered particle (TLP) formation was measured by preparing lysates from tsE-infected cells that were transfected with IR siRNA, g8D siRNA, or g8D siRNA and an NSP2a* expression vector and maintained in media containing 35S-labeled amino acids. Virions in the lysates were detected by CsCl centrifugation, followed by electrophoresis of the gradient fractions (Fig. 6). Virions were identified based on their density (ρ = 1.36 g/ml) and protein content, notably, the inner, intermediate, and outer shell proteins VP2, VP6, and VP7, respectively. Fractions containing virions were pooled, and the yield of infectious virus was quantified by IFF assay using anti-VP6 antsiera. The analysis revealed that NSP2a* expression in the g8D siRNA-transfected cells promoted virion assembly to levels approximately four- to sixfold higher than that of similarly treated cells not producing NSP2a* (Fig. 6). Thus, NSP2* expression rescued defects in viroplasm formation, genome replication, and virion assembly in tsE-infected cells containing g8D siRNA, findings validating the potential of the complementation system as a tool for dissecting the structure and function of NSP2.
FIG. 6.
Virus assembly in NSP2*-complemented cells. MA104 cells were transfected with either IR (A) or g8D siRNA (B and C) and with a vector expressing NSP2* (C) and then infected with vTF7.3 and the tsE strain. Cells were maintained in the presence of 35S-labeled amino acids and harvested ∼18 h p.i. Virus particles pelleted (p) from cell lysates were analyzed by CsCl centrifugation. Gradient fractions were analyzed for density and for protein content by electrophoresis. The titer of the virus in the fractions containing TLPs (*) was determined by IFF assay.
NTPase activity of NSP2 is required for dsRNA synthesis.
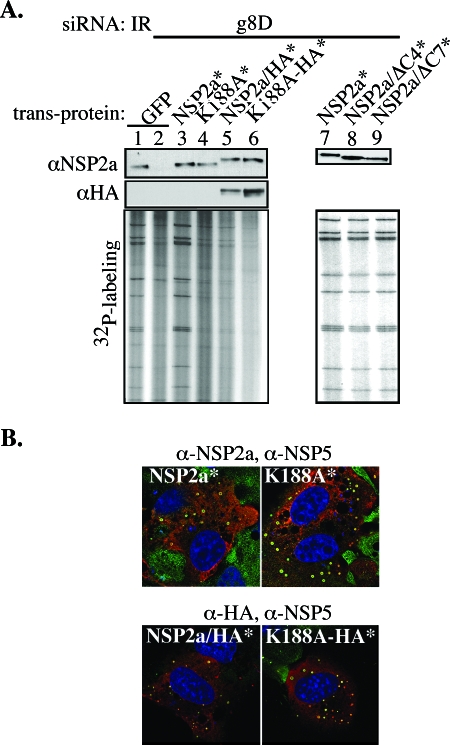
NSP2 has been implicated in rotavirus replication and packaging (6, 27, 29), but the contribution of the NTPase activity of NSP2 to these processes is uncertain. The enzymatic activity could be anticipated to have an essential function, given the conservation of the HIT-like motif in the NSP2a and NSP2c octamer and the retention of the activity. To examine this possibility, the abilities of two mutant forms of NSP2a (K188A and H225A) possessing little or no NTPase activity were assayed in the complementation system for the ability to support genome replication and viroplasm formation. The NTPase-defective phenotype of K188A* and H225A* results from mutation of conserved residues that make up the HIT-like motif (4). The sedimentation properties of the bacterially expressed K188A and H225A were indistinguishable from that of NSP2a (4), indicating that the mutant forms of NSP2 retained the octamer conformation. The complementation assay showed that K188A* failed to promote dsRNA synthesis above the level detected in control cells lacking transiently expressed NSP2 (Fig. 7A, lanes 2 and 4). The level of dsRNA synthesis with K188A* was less than that in cells containing NSP2a* (Fig. 7A, lane 3). Thus, the NTPase-defective phenotype was correlated with an inability of NSP2a* to support genome replication, indicating that the NTPase activity of NSP2 is critical for virus replication. Similar results were obtained using H225A* in the complementation system (data not shown).
FIG. 7.
Effect of NSP2 mutation on genome replication. MA104 cells were transfected with an siRNA (IR or g8D) and a transcription vector expressing GFP or wt or mutant NSP2 and then infected with vTF7.3 and the tsE strain and maintained in medium containing no label or 32Pi. At 12 h p.i., the infected cells were analyzed for protein expression, dsRNA synthesis, and viroplasm formation. (A) NSP2a and HA fused to NSP2a were detected by Western blot assay, and 32P-labeled dsRNA samples were analyzed by electrophoresis on 12% polyacrylamide gels and autoradiography. (B) Viroplasm formation in cells producing the indicated vector-derived NSP2a was monitored by IF assay using primary antibodies (anti-NSP2a [αNSP2a] and anti-HA [αHA]) for the detection of NSP2a* (green) and NSP5 (red) in the upper panel and for the detection of the HA-tagged NSP2a* (red) and NSP5 (green) in the lower panel. Colocalization of signals is indicated by yellow, and cell nuclei were visualized by DAPI.
To determine whether K188A* and H225A* were defective in supporting viroplasm formation, cells expressing these proteins in complementation assays were analyzed by IF. As shown in Fig. 7B, K188A* supported the formation of numerous large viroplasms, like those formed in cells expressing NSP2a*. Similar viroplasms were detected in cells expressing H225A* (data not shown). The fact that K188A* failed to support dsRNA synthesis, despite supporting viroplasm formation, suggests that the NTPase activity of NSP2 contributes to virus replication at a step independent of viroplasm formation. These results also show that the K188A mutation does not prevent NSP2a from interacting with NSP5a, since viroplasm formation is dependent on this event. Our findings are consistent with the results of previous coexpression assays, which showed that both the K188A and H225A mutant forms can interact with NSP5 to form VLS in uninfected cells (4).
C-terminal HA tag interferes with NSP2 function in replication.
To distinguish between virus-encoded NSP2 and transiently expressed NSP2a* and K188A* in IF assays, and thereby confirm that NSP2a* and K188A* promoted viroplasm formation in the complementation system, a C-terminal HA tag (YPYDVPDYA) was added to these proteins. The resulting products, NSP2a/HA* and K188A-HA*, when transiently expressed in the complementation system, supported viroplasm formation (Fig. 7B). Hence, addition of short C-terminal sequences such as the HA tag does not interfere with the capacity of NSP2a to support viroplasm formation. This finding is consistent with earlier results showing that NSP2a linked at its C terminus to GFP supported VLS formation when transiently coexpressed with NSP5a (10). Thus, the addition of extra residues to the C terminus of NSP2 does not prevent the necessary interaction between the protein and NSP5a required for viroplasm formation.
The functionality of NSP2a/HA* and K188A-HA* was further evaluated by analyzing their capacity to support dsRNA synthesis in the complementation system. In agreement with the results described above with the K188A* mutant (Fig. 7A), the tagged version of the protein, K188A-HA*, failed to stimulate dsRNA synthesis above levels detected in cells lacking NSP2a* (lane 6). Surprisingly, the HA-tagged form of NSP2a* (lane 5), unlike the untagged form (lane 3), also failed to support dsRNA synthesis above background levels (lane 2). These results indicate that although addition of HA tag to NSP2a* did not affect its function in viroplasm formation, the modification did render the protein biologically inactive based on its failure to support genome replication.
The C-terminal tail of NSP2 is dispensable for replication.
The rationale for adding the HA tag to the C terminus of NSP2a was, in part, based on the observation that the C-terminal amino acids 314 to 317 of NSP2 and the fused six-His tag were not detectable in the NSP2a (and NSP2c) crystal structure and therefore were assumed to be disordered (18). However, despite its disordered status, the complementation assay suggested that the addition of the HA tag to the C terminus interfered with an undefined, but important biological function of the protein. To further investigate the importance of the C terminus, transcription vectors were constructed that encoded NSP2a* with C-terminal deletions of 4 (ΔC4*) or 7 (ΔC7*) amino acids. These modified proteins were compared with NSP2a* for the ability to support dsRNA synthesis in complementation assays. As shown in Fig. 7A (lanes 7 to 9), dsRNA levels in cells expressing ΔC4* or ΔC7* were like those in cells expressing NSP2a*. Thus, the C terminus of NSP2a (311 to 317) has no structure or function that is essential for rotavirus genome replication. Our results showing that the terminal amino acids are not required for replication suggest that the HA tag interfered with NSP2 function not by perturbing a direct role of the C terminus in dsRNA synthesis, but more likely by obstructing other undefined functional domains of the octamer.
Group C NSP2 fails to substitute for group A NSP2.
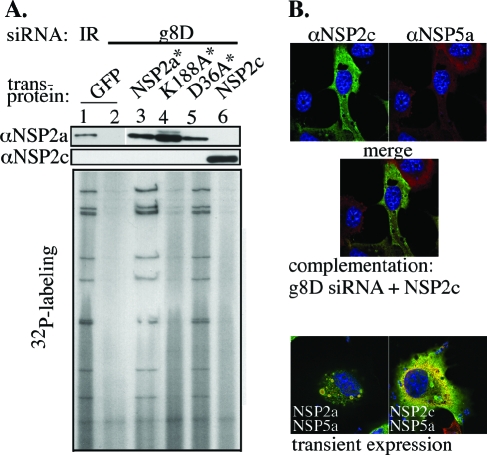
The similarity of the NSP2a and NSP2c octamers both in their structure and activity (NTPase and RNA binding) suggests that these proteins are capable of substituting for one another in the viral life cycle. To address this possibility, we used the complementation system to test whether transiently expressed NSP2c*, in place of NSP2a*, could support genome replication and viroplasm formation in tsE-infected cells. As shown in Fig. 8A, little or no dsRNA was produced in cells containing NSP2c (lane 6), similar to results with K188A* (lane 4). On the other hand, cells containing NSP2a* (lane 3) synthesized dsRNA to levels similar to that of cells containing virion-encoded NSP2a (lane 1). NSP2a/D36A*, a form of NSP2a with a nondisruptive mutation mapping to the tetramer-tetramer interface of the octamer and positioned away from any putative ligand-binding site, also supported dsRNA to levels comparable to that of NSP2a*. Taken together, these results show that despite structural similarity, the NSP2c octamer cannot functionally replace NSP2a in supporting replication of the tsE strain, a group A rotavirus.
FIG. 8.
Failure of NSP2c to support group A rotavirus replication. MA104 cells were transfected with an siRNA (IR or g8D) and a transcription vector expressing GFP, wt or mutant NSP2a*, or NSP2c, and then infected with vTF7.3 and the tsE strain. (A) At 12 h p.i., the infected cells were analyzed for protein expression by Western blot assay and 32P-labeled viral dsRNA was analyzed by 12% polyacrylamide gel electrophoresis and autoradiography. αNSP2a, anti-NSP2a; αNSP2c, anti-NSP2c; αNSP5a, anti-NSP5a. (B) Formation of viroplasms in g8D-treated tsE strain-infected MA104 cells expressing NSP2c and VLS in MA104 cells transiently expressing either NSP2c and NSP5a or NSP2a and NSP5a was monitored at 12 h p.i. by IF assay (NSP2, green; NSP5, red; colocalization, yellow; nuclei, blue).
The complementation system showed that NSP2c* not only failed to support dsRNA synthesis in tsE-infected cells, but based on IF analysis, also failed to nucleate viroplasm formation with NSP5a (Fig. 8B). Instead, NSP2c* and tsE-encoded NSP5a were diffusely distributed throughout the cytoplasm. Similarly, transient coexpression of NSP2c and NSP5a in uninfected cells did not generate VLS. These data indicate that NSP2c cannot interact with NSP5a for VLS formation. Because both the NSP2a and NSP2c octamers share similar nonspecific RNA-binding and NTPase activities, it seems doubtful that either of these has something to do with the failure of NSP2c to support viroplasm formation. Instead, given the obvious difference in the electrostatic surface potential of the NSP2a and NSP2c octamers, it seems more reasonable to predict that the unique surface charge on the Bristol species prevented it from interacting with NSP5a in such a way as to form the NSP2-NSP5 complexes required for viroplasm assembly.
DISCUSSION
In this study, we report that NSP2a and NSP2c display a considerable degree of structural conservation, both in their fold as monomers and in the arrangement of NSP2a and NSP2c monomers within octamers. NSP2a and NSP2c also retain a common HIT-like fold and use a nearly identical set of residues to create the HIT-like signature motif that is associated with the NTPase activity of the protein. Although sharing a similar architecture, assays performed with an NSP2-dependent complementation system show that NSP2c cannot functionally replace NSP2a, as it fails to rescue genome replication in NSP2a-depleted cells infected with a group A virus. This failure appears connected to the inability of NSP2c to interact appropriately with NSP5a to form viroplasms in rotavirus-infected cells. This is also consistent with the observation that coexpression of NSP2c and NSP5a fails to stimulate VLS formation. Likewise, the coexpression of NSP2a and NSP5c does not result in VLS formation (unpublished data). The molecular interactions that are required to generate NSP2-NSP5 complexes have not been defined; however, considering that NSP2a (pI ∼9.02) is more basic than NSP2c (pI ∼8.6) and that NSP5c (pI ∼4.9) is significantly more acidic than NSP5a (pI ∼6.7), it is likely that charge complementarity plays an important role. GRASP (graphical representation of surface potential) analysis shows that the distribution of charged residues on the NSP2c octamer is quite distinct from that on the NSP2a octamer (Fig. 3C and D). The different arrangement of charges on the NSP2c octamer may make the NSP2c incompatible for the necessary interactions with NSP5a, required to generate the complexes that serve as building blocks in viroplasm formation. However, we cannot exclude the possibility that one of the minor structural differences observed between the NSP2a and NSP2c octamers (e.g., 170-to-185 loop) accounts for their functional incompatibility.
Previous studies analyzing the formation of TLPs by coexpression of different combinations of group A and C capsid proteins have shown that these proteins can functionally substitute for each other in assembly, thereby allowing the production of chimeric particles (22). The fact that such chimeric particles can be generated indicates that surface residues on the group A and C capsid proteins that drive the necessary protein-protein interactions required for particle assembly are conserved. Thus, while some interactions between group A and C proteins are possible, such as those involving capsid proteins, other interactions are not, such as those for the nonstructural proteins NSP2 and NSP5. The inability of NSP2 and NSP5 of different rotavirus groups to collaborate in forming chimeric viroplasms may preclude the establishment of viral factories in coinfected cells with the capacity to replicate and package, simultaneously, genomes belonging to two different groups. Without such chimeric viroplasms, formation of reassortants by the mixed packaging of newly replicated dsRNA into progeny cores is, presumably, not possible. Indeed, given the essential contribution of viroplasms to packaging and replication of the viral genome, an inability of the NSP2 and NSP5 proteins of different rotavirus groups to function together in viroplasm assembly would impose a dominating insurmountable restriction on reassortment, one that could be not be overcome even if all other viral proteins were fully compatible in their functions.
To establish a biologically relevant assay system for the study of NSP2 function, we infected cells with a mutant virus containing a ts lesion mapping to the NSP2 gene (tsE) and used an siRNA to induce the degradation of the NSP2 gene in the tsE-infected cells. This combination at the nonpermissive temperature resulted in little or no NSP2 expression, dsRNA replication, and viroplasm formation in the cell, establishing the basis for the creation of an NSP2-dependent complementation system. The success in applying the NSP2-dependent complementation system to the analysis of NSP2 mutants in the viral life cycle suggests that combinations of other SA11 ts mutants and siRNAs may be similarly used for defining the functions of other rotavirus proteins: VP1 (tsC), VP2 (tsF), VP3 (tsB), VP4 (tsA), and VP6 (tsG) (14). Interestingly, although providing NSP2 in trans in the NSP2-dependent complementation system stimulated dsRNA synthesis, viroplasm formation, and TLP assembly, the levels of these events were not as high as those that occur in wt SA11-infected cells. The explanation for this is not known, but may, to some extent, reflect the technical impossibility of successfully introducing two viruses, one siRNA, and one plasmid into all the cells used in these experiments. Others have also encountered difficulties in creating protein-dependent complementation systems that can fully restore levels of virus replication. An investigation of this limitation for a complementation system developed for flock house virus indicates that the failure of the protein provided in trans to support replication events may be a reflection of not just how much protein is made, but also where the protein is made in the cell (36). That is, fully restoring levels of virus replication may require the trans protein to be produced in a manner that is spatially coordinated with the site in which the protein is utilized. In the case of the NSP2-dependent complementation system, it may be reasoned that NSP2 in trans does not support rotavirus replication with maximum efficiency because much of the protein is made distantly from the viroplasms in which the protein is needed, instead of being made in polysomal fields adjoining viroplasms.
By using the complementation assay, we provide the first evidence of the dependency of rotavirus dsRNA synthesis on nucleotide hydrolysis by NSP2. The importance of this finding is underscored by the uniqueness of this activity. Unlike other viral NTPases, NSP2 does not belong to the large superfamily of viral NTPases/helicases that carry one or more of the characteristic Walker motifs (15, 16, 20). Instead, based on structural homology, a HIT-like motif was identified as the putative site for nucleotide binding and hydrolysis in NSP2 (18) and later confirmed by site-directed mutagenesis (4). This was the first report of a viral NTPase that shared features with a prototype member in the large and ubiquitous nucleotide-binding HIT family of proteins. Another uncommon feature of the NTPase activity of NSP2 is that it is not stimulated by ssRNA and does not drive a helicase-like function. However, NSP2 does possess an NTP- and Mg2+-independent helix-destabilizing activity (35), a passive function that is purely stoichiometric and not kinetic and that is often found associated with viral ssDNA binding proteins (1, 25, 30, 33).
Most viral NTPases are believed to be involved in duplex unwinding during transcription, packaging, replication, translation, recombination, or repair (15). In this study, we show that genome replication does not proceed without nucleotide hydrolysis by NSP2 in rotavirus-infected cells despite the formation of viroplasm. A simple interpretation of this finding is that the NTPase activity of NSP2 comes into play after the establishment of subcellular sites for replication and assembly, possibly during the formation of replication intermediates. Because nucleotide binding occurs in the interdomain cleft region in each NSP2 monomer (4) and induces conformation shifts in the NSP2 octamer from a more relaxed to a compact conformation (31), one would predict that nucleotide-induced conformational shifts transduce structural changes in the neighboring electropositive grooves of the NSP2 octamer. These deep grooves are sites where the ssRNA is believed to wrap around the periphery of the octamer (18). Hence, nucleotide binding and hydrolysis could regulate the interaction of NSP2 with ssRNA. The recent finding that an ssRNA molecule with a 5′ γ-phosphate is a better substrate for the hydrolytic activity of NSP2 than an NTP and involves the same HIT-like active site (R. V. Carpio et al., unpublished observations) further supports the notion that the hydrolytic activity of NSP2 may regulate the ssRNA binding activity, or vice versa, possibly, during genome packaging or the initiation of dsRNA synthesis.
Supplementary Material
Acknowledgments
We appreciate the contribution of Bristol gene 9 cDNA by Ian Clark (Southampton Hospital, United Kingdom), the tsE(1400) strain by Frank Ramig (Baylor College of Medicine, Houston, Tex.), and mouse anti-NSP5 monoclonal antibody 158G37 by Didier Poncet (CNRS-INRA, France).
This work was supported by the Intramural Research Program of NIAID, NIH, to J.T.P. and grants from the NIH (AI36040) and R. Welch Foundation to B.V.V.P.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Boehmer, P. E., and I. R. Lehman. 1993. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J. Virol. 67:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, C., P. Bieganowski, H. C. Pace, and K. Huebner. 1999. The histidine triad superfamily of nucleotide-binding proteins. J. Cell Physiol. 181:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 4.Carpio, R. V., F. D. Gonzalez-Nilo, H. Jayaram, E. Spencer, B. V. Prasad, J. T. Patton, and Z. F. Taraporewala. 2004. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J. Biol. Chem. 279:10624-10633. [DOI] [PubMed] [Google Scholar]
- 5.Caul, E. O., C. R. Ashley, J. M. Darville, and J. C. Bridger. 1990. Group C rotavirus associated with fatal enteritis in a family outbreak. J. Med. Virol. 30:201-205. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D., J. L. Gombold, and R. F. Ramig. 1990. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology 178:143-151. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., P. R. Lambden, J. Lau, E. O. Caul, and I. N. Clark. 2002. Human group C rotavirus: completion of the genome sequences and gene coding assignments of a non-cultivatable rotavirus. Virus Res. 83:179-187. [DOI] [PubMed] [Google Scholar]
- 8.Chizhikov, V., and J. T. Patton. 2000. A four-nucleotide translation enhancer in the 3′-terminal consensus sequence of the nonpolyadenylated mRNAs of rotavirus. RNA 6:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.Eichwald, C., J. F. Rodriguez, and O. R. Burrone. 2004. Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J. Gen. Virol. 85:625-634. [DOI] [PubMed] [Google Scholar]
- 11.Estes, M. K. 2001. Rotaviruses and their replication, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. R. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gombold, J. L., and R. F. Ramig. 1987. Assignment of simian rotavirus SA11 temperature-sensitive mutant groups A, C, F, and G to genome segments. Virology 161:463-473. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya, A. E., E. V. Koonin, A. P. Donschenko, and V. M. Blinov. 1988. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 235:16-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honig, B., and A. Nicholls. 1995. Classical electrostatics in biology and chemistry. Science. 268:1144-1149. [DOI] [PubMed] [Google Scholar]
- 18.Jayaram, H., Z. Taraporewala, J. T. Patton, and B. V. Prasad. 2002. Rotavirus protein involved in genome replication and packaging exhibits a HIT-like fold. Nature 417:311-315. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 20.Kadaré, G., and A.-L. Haenni. 1997. Virus-encoded RNA helicases. J. Virol. 71:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapikian, A. 2001. A rotavirus vaccine for prevention of severe diarrhoea of infants and young children: development, utilization and withdrawal. Novartis Found. Symp. 238:153-171. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y., K. O. Chang, W. Y. Kim, and L. J. Saif. 2002. Production of hybrid double- or triple-layered virus-like particles of group A and C rotaviruses using a baculovirus expression system. Virology 302:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Komoto, S., J. Sasaki, and K. Taniguchi. 2006. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA 103:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. Sect. D Biol. Crystallogr. 61:458-464. [DOI] [PubMed] [Google Scholar]
- 25.Monaghan, A., A. Webster, and R. T. Hay. 1994. Adenovirus DNA binding protein: helix destabilising properties. Nucleic Acids Res. 22:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 27.Patton, J. T., and C. O. Gallegos. 1988. Structure and protein composition of the rotavirus replicase particle. Virology 166:358-365. [DOI] [PubMed] [Google Scholar]
- 28.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277:217-225. [DOI] [PubMed] [Google Scholar]
- 29.Ramig, R. F., and B. L. Petrie. 1984. Characterization of temperature-sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J. Virol. 49:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rochester, S. C., and P. Traktman. 1998. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J. Virol. 72:2917-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck, P., Z. Taraporewala, P. McPhie, and J. T. Patton. 2001. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. 276:9679-9687. [DOI] [PubMed] [Google Scholar]
- 32.Silvestri, L. S., Z. F. Taraporewala, and J. T. Patton. 2004. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J. Virol. 78:7763-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soengas, M. S., C. Gutierrez, and M. Salas. 1995. Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J. Mol. Biol. 253:517-529. [DOI] [PubMed] [Google Scholar]
- 34.Taraporewala, Z., D. Chen, and J. T. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taraporewala, Z. F., and J. T. Patton. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venter, P. A., N. K. Krishna, and A. Schneemann. 2005. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 79:6239-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.