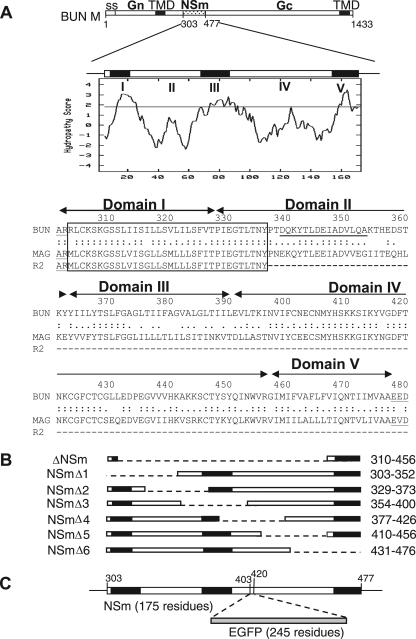
FIG. 1.
The BUNV M RNA segment and the encoded precursor polyprotein. (A) The gene layout of the M segment is shown at the top, with positions of amino acid residues marking protein boundaries indicated. ss, signal peptide; TMD, transmembrane domain. Below, the Kyte-Doolittle hydropathy plot and predicted domain structure of NSm are shown. Domains I to V were suggested by the program TMHMM (21). The amino acid alignment of the NSm proteins of BUNV, Maguari virus, and its mutant R2 are shown, with the conserved N-terminal region boxed. The peptide sequence used to raise the anti-NSm antibody is underlined. (B) Schematic of NSm deletion mutants. The regions deleted are indicated by the dashed line, and the residues deleted are indicated at the right. (C) Insertion of EGFP open reading frame into NSm. The EGFP coding sequence was cloned into the M segment cDNA at artificial SacI restriction enzyme sites created at codons 403 and 420 in NSm.