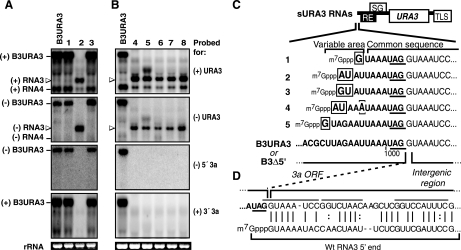
FIG. 4.
Sample Northern blot analyses of URA3-expressing BMV RNA replicons in cells that acquired a Ura+ phenotype. Positive- and negative-strand RNAs were detected with 32P-labeled probes as indicated in Fig. 2. wt B3URA3 was included as a size marker. (A) Examples of the two classes of RNA progeny detected in Ura+ cells obtained after transcription of both B3Δ3′ and B3Δ5′. Homologous intermolecular recombination products (lanes 1 and 3) matched the size of B3URA3 and were detected with both parental RNA-specific probes. sURA3 RNAs (lane 2) were only slightly longer than RNA4 and failed to hybridize to probes targeting 3a sequences. Open arrowheads point to sURA3 of both positive and negative polarity detected with URA3 probes. (B) sURA3 RNAs obtained after transcription of B3Δ5′ alone in the absence of pB3Δ3′. (C) Nucleotide sequences at the 5′ end of sURA3 RNAs aligned with the 3′ end of the 3a ORF in B3URA3 and B3Δ5′. The 3a stop codon is underlined. The in vivo-generated 5′ ends consisted of the last 7 to 11 nucleotides from the 3a ORF and all downstream sequences of B3Δ5′. An additional G, GU, or AU was present at the extreme 5′ end (indicated by a box). For clone number 4, nucleotide 5 (bracketed) was a U-to-A substitution. Sequences derived from the 3a ORF are shown in boldface type. (D) 5′ ends of sURA3 RNAs aligned with the 5′ end of wt RNA3.