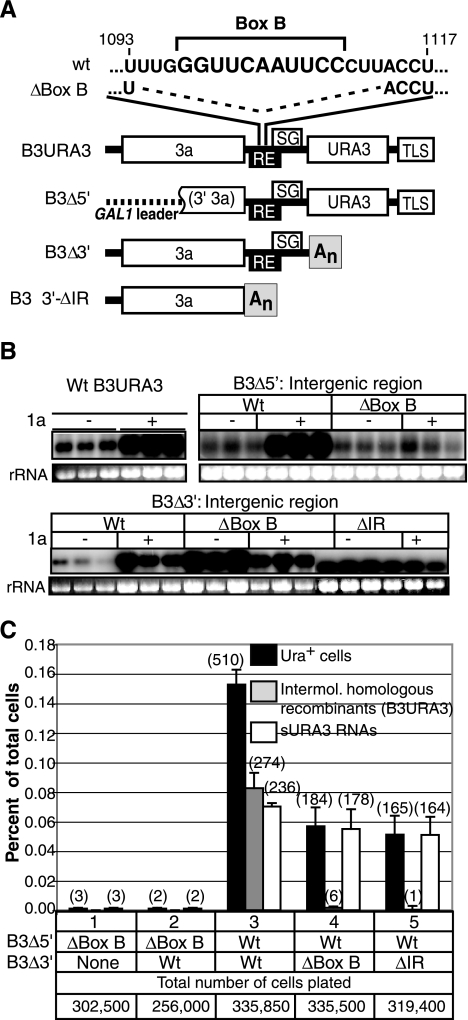
FIG. 7.
(A) wt B3URA3 and its derivatives (B3Δ5′, B3Δ3′, and B3Δ3′ΔIR) are shown. Within the RE, the nucleotide sequence of box B is indicated, along with the box B deletion (ΔBox B). The intergenic region was removed from B3Δ3′ to generate B3Δ3′ΔIR. (B) RNA accumulation in the presence (+) or absence (−) of BMV replication protein 1a after transcription for 72 h. For each construct, six independent cultures were analyzed: three in the presence and three in the absence of 1a. B3URA3 and B3Δ5′ were transcribed from the GAL1 promoter, while B3Δ3′ and its derivatives were transcribed from the CUP1 promoter. Three micrograms of total RNA was fractionated in 1% denaturing agarose gels. Positive-strand RNA sequences were detected with a 32P-labeled 3′3a common RNA probe (Fig. 1B). B3URA3 was used as a size marker and positive control. Ethidium bromide staining of 18S rRNA in the same samples is indicated at the bottom of each panel. (C) Frequency of Ura+ cells and RNA recom binants obtained after transcription of parental RNAs with or without a functional RE in 1a2a+ yeast. The histogram shows the averages and standard errors of results from three independent repetitions. Black bars represent the percentages of cells that acquired the Ura+ phenotype and harbored either an intermolecular homologous RNA recombinant (gray bar) or an sURA3 RNA (white bars). Numbers in parentheses indicate the total number of cases observed in three independent repetitions of the experiment. The corresponding total number of cells plated is indicated.