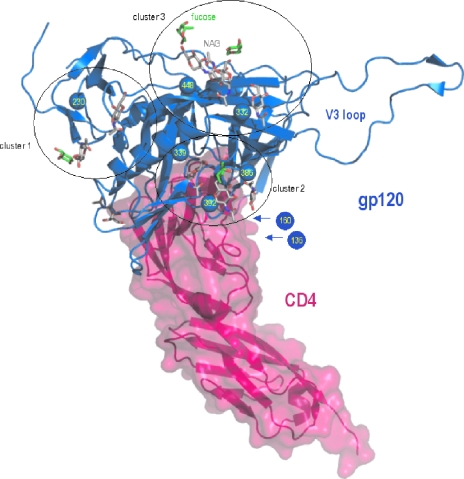
FIG. 3.
Ribbon representation of the HIV-1 gp120 envelope glycoprotein (blue) bound to CD4 receptor (pink), as found in a composite of X-ray crystal structures with Protein Data Bank identification codes 1GC1 (19, 32) and 2B4C (18). Sugar moieties are displayed as sticks, with C atoms colored white for N-acetyl-d-glucosamine and green for fucose. The Cbeta atoms of the glycosylated N residues that are mutated in the presence of CV-N are displayed as spheres and have been labeled. The PyMOL molecular graphics system (http://www.pymol.org; DeLano Scientific LLC, San Carlos, CA) was used for visualization and picture creation. Clustering was done manually on the basis of proximity of the glycosylated sites. Residues 127 to 192 making up loops V1 and V2 are absent in the crystal structures of gp120. Therefore, the approximate positions of the N-136 and N-160 residues are indicated by arrows.