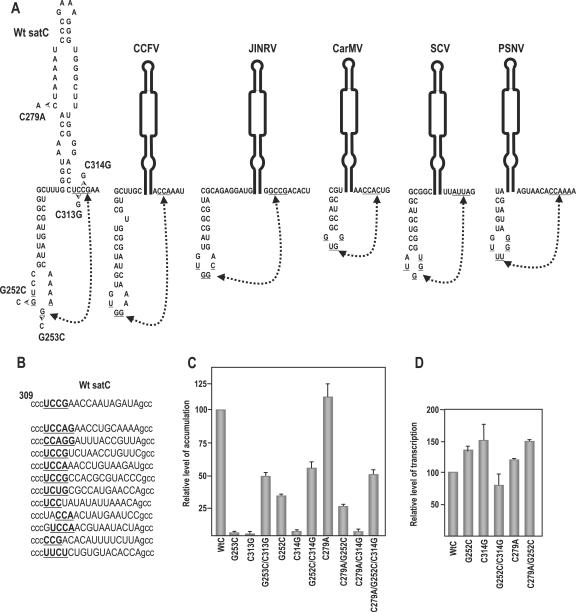
FIG. 5.
Interaction between positions 251 and 254 and positions 312 and 315 is important for satC accumulation in vivo. (A) Phylogenetically inferred structure of the H4b/H5 region. Potential base pairing between underlined residues is denoted by connected arrowheads. Mutations generated in putative base-paired partners are shown. Location of alteration C279A, which stabilizes H5 by pairing the lower positions in the LSL, is shown. Also presented are putative pairings between H4b loop sequences and H5 flanking sequence that can occur in other carmoviruses. JINRV, Japanese iris necrosis virus; CarMV, Carnation mottle virus; SCV, Saguaro cactus virus; PSNV, Pea stem necrosis virus. While the interaction is shown in the active form of the 3′ region of satC for convenience, this does not imply that the interaction exists in this structural configuration. (B) Sequences found in satC accumulating in plants after in vivo functional selection. satC containing randomized sequence in positions 312 to 327 (uppercase) were inoculated onto turnip plants with TCV genomic RNA, and functional satC from six plants was recovered 3 weeks later. The top sequence is wt satC. Underlined, boldface bases maintain possible pairing with H4b loop sequences. (C) Accumulation of satC with various mutations in protoplasts at 40 hpi. Mutations are described above for panel A. Data from at least three independent experiments were normalized to wt satC levels, arbitrarily assigned a value of 100. (D) In vitro transcription of wt satC and mutant transcripts by use of purified recombinant TCV p88. Data are from three independent experiments. Error bars denote standard deviations.