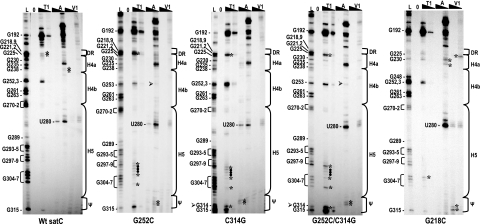
FIG. 6.
Disrupting either pseudoknot partner results in nearly identical structural changes in the preactive structure of satC in vitro. Names of the transcripts used for solution structure analysis are given below each autoradiograph. Transcripts were subjected to limited cleavage with two concentrations each of RNase T1 (T1), RNase A (A), or RNase V1 (V1). L, RNase T1 ladder; 0, no added enzymes. High and low concentrations of each enzyme are indicated by the filled triangles above the lanes. Positions of guanylates determined from the T1 ladder reactions are given at left, and the position of U280 is also shown. Locations of sequences within the phylogenetically inferred hairpins and flanking regions are denoted by brackets at right. Ψ denotes positions 312 to 315 (UCCG) involved in Ψ2. Arrowheads denote new guanylates or cytidylates in constructs containing C314G and G252C, respectively. Asterisks denote enzyme-specific cleavages that differ between mutants and wt satC. Filled circles denote nonspecific pyrimidine cleavages by RNase T1 using mutant transcripts.