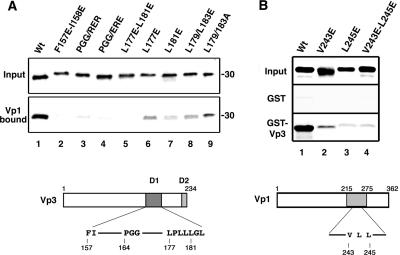
FIG. 5.
In vitro interaction of point mutant Vp1s and Vp3s. (A) 35S-labeled Vp3s were reacted with the Vp1ΔC58 pentamer, and the bound proteins were separated by SDS-PAGE followed by fluorography. The upper panel shows one-fifth of the Vp3 protein used for the reaction (Input). The lower panel shows one-half of the protein retrieved from the Vp1ΔC58-bound beads (Vp1 bound). A schematic diagram of Vp3 with locations for mutations within D1 marked is shown at the bottom. The wild type (Wt) and the following mutant Vp3 proteins were examined: lane 1, wild type; lane 2, F157E-158E; lane 3, PGG/RER; lane 4, PGG/ERE; lane 5, L177E-L181E; lane 6, L177E; lane 7, L181E; lane 8, L179E/L183E; and lane 9, L179A/L183A. (B) 35S-labeled Vp1s were reacted with either GST or GST-Vp3, and the bound proteins were retrieved by glutathione-Sepharose beads. The bound Vp1 was visualized by SDS-PAGE followed by fluorography. The upper panel shows one-fifth of the input count used for reaction (Input), the middle panel shows one-half of the GST-bound Vp1s (GST), and the bottom panel shows one-half of the GST-Vp3-bound Vp1s (GST-VP3). Compared to what was found for the wild type, 53%, 14%, and 9% of the Vp1 V243E, L245E, and V243E-L245E mutants, respectively, were retrieved from the GST-Vp3-bound beads. A schematic diagram of Vp1 with mutations marked is shown at the bottom. The following 35S-labeled Vp1s were used: lane 1, wild type; lane 2, V243E; lane 3, L245E; and lane 4, V243E-L245E.