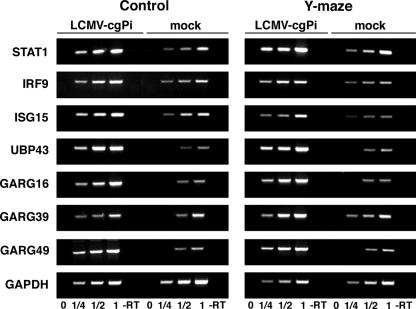
FIG. 5.
Validation of changes in gene expression of selected candidate genes in LCMV-cgPi mice. Total RNA was isolated from hippocampus tissue, and contaminant DNA was removed. A reverse transcription (RT) reaction was performed using 5 μg RNA and random hexamer primers, and specific PCR fragments were amplified using specific primers for the candidate genes indicated and the housekeeping gene GAPDH. For semiquantitative analysis the linear range of PCR product-template was determined by serial dilution of the RT products obtained with the mock-infected samples. To detect quantitative differences in mRNA concentration of the candidate genes between LCMV-cgPi and control samples, PCR was performed on identical RT product dilutions within the linear range of PCR product-template. Results for mock-infected controls and LCMV-cgPi animals either from the control group (control) or exposed to the Y-maze learning paradigm (Y-maze) are shown. A control reaction without RT (−RT) is indicated.