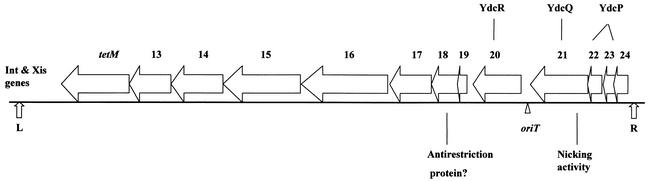
FIG. 4.
Map of the conjugative transposon Tn916 with open reading frames thought to be related to conjugation. The genes related to excision of Tn916, int and xis, are shown near the left end of the transposon. tetM is the inducible tetracycline resistance determinant of the transposon. Orf13 and Orf14 have homologues (near identity) within Tn5397, a conjugative transposon in Clostridium difficile. Orf15 and Orf16 each have homologues on both pAD1 and pAM373. Orf18 has similarity to the bacterial antirestriction proteins, Ard of plasmid Coll-b-P9 and ArdA of pKM101 (72). Orf20 is a homologue of the hypothetical YdcR in B. subtilis; Orf22 and Orf23, which have some similarity to each other, are both homologues of the hypothetical YdcP. Orf21 has nonspecific nicking activity that is likely to be related to an origin-involved transfer event (44). Orf21 has strong homology to a B. subtilis hypothetical protein (YdcQ), and an internal segment resembles the FtsK/SpoIIIE family (14). Modified from reference 44.