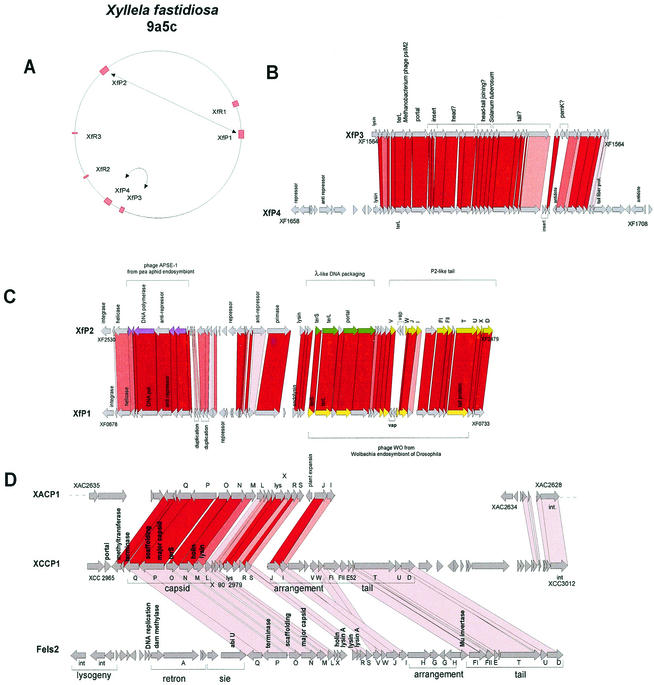
FIG. 13.
Prophages from bacterial plant pathogens. (A) Prophage content of X. fastidiosa strain 9a5c. Prophages linked by high DNA sequence identity are connected by double-arrowed lines. (B) Alignment of the X. fastidiosa prophages XfP3 and XfP4. The degree of protein sequence identity is reflected by different intensities of red shading. Suspected gene functions are indicated. (C) Alignment of the X. fastidiosa prophages XfP1 and XfP2. Selected genes are annotated. Genes with protein sequence similarity to phage APSE-1 from an endosymbiont of a pea aphid are marked in violet, and those with similarity to phage WO from the Drosophila endosymbiont Wolbachia are marked in yellow. Genes corresponding to lambda-like DNA-packaging genes are in dark green, while tail genes resembling P2 phage proteins are in light green. The gene numbers for the first and last genes of the prophages are indicated to facilitate orientation on the Xylella genome. Candidates for virulence-associated genes are marked with vap. Prophage genes with protein sequence identity are linked by shading. The different shades of red indicate >90%, >80%, >70%, and any protein sequence identity. (D) Alignment of the X. campestris prophage XCCP1 (center) with X. axonopodis prophage XACP1 (top) and Salmonella prophage Fels-2 (bottom). Note that the leftmost XACP1 genes are virtually translocated and inverted to allow an optimal alignment with the XCCP1 gene map.