Figure 6.
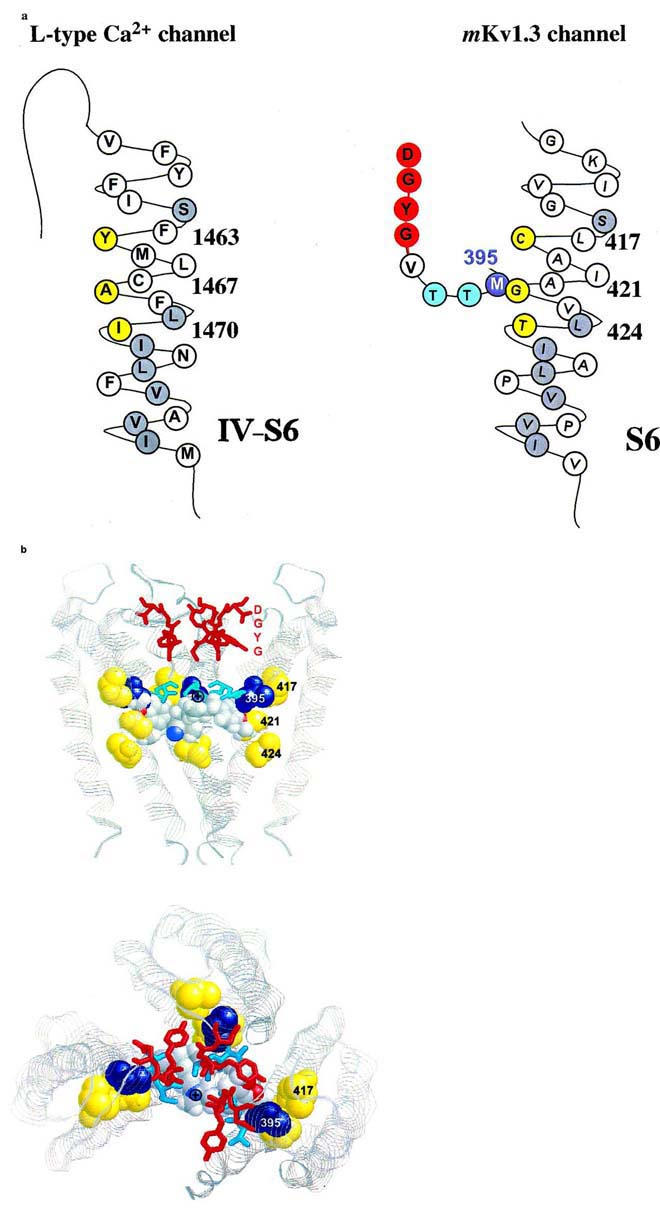
Proposed interaction of amino acid residues with verapamil. (A) Amino acid sequence of the S6 transmembrane segment of domain VI of the L-type Ca2+ channel (left) and of the deep-pore and the S6 transmembrane segment of the voltage-gated Kv1.3 channel (right) are shown either in their predicted helical conformation (Hockerman et al., 1997) or derived from the crystal structure of the KcsA channels (Doyle et al., 1998), respectively. The amino acids of the postulated phenylalkylamine binding site are highlighted in yellow. The homologous amino acids identical to S6 residues in Kv1.3 are highlighted in gray. The putative binding residue M395 in the deep-pore of Kv1.3 is highlighted in blue. (B) Proposed model of the verapamil interaction with the internal pore region. Shown are three subunits of the KcsA channel as ribbons in the conformation derived from the crystal structure. The homologous amino acids at position 417, 421 and 424 in the S6-segment of Kv1.3, corresponding to the L-type Ca2+ channel phenylalkylamine binding sites, are shown in yellow (spacefill), the homologous internal residues to M395 are shown in blue (spacefill). The amino acids of the GYGD-motif (red) and the residues corresponding to T396 and T397 (cyan) are shown as sticks. Verapamil is shown in spacefill with the usual atom colors. Top: side view of the KcsA channel with the proposed orientation of verapamil. The phenyl rings are located in a hydrophobic environment in closest proximity to the residues homologous to M395 and G421. The protonated nitrogen (⊕) is facing up towards the narrow part of the inner pore. Bottom: view from above into the channel pore. The protonated nitrogen of verapamil is visible in the center of the pore facing towards the negative environment of the GYGD-motif.
