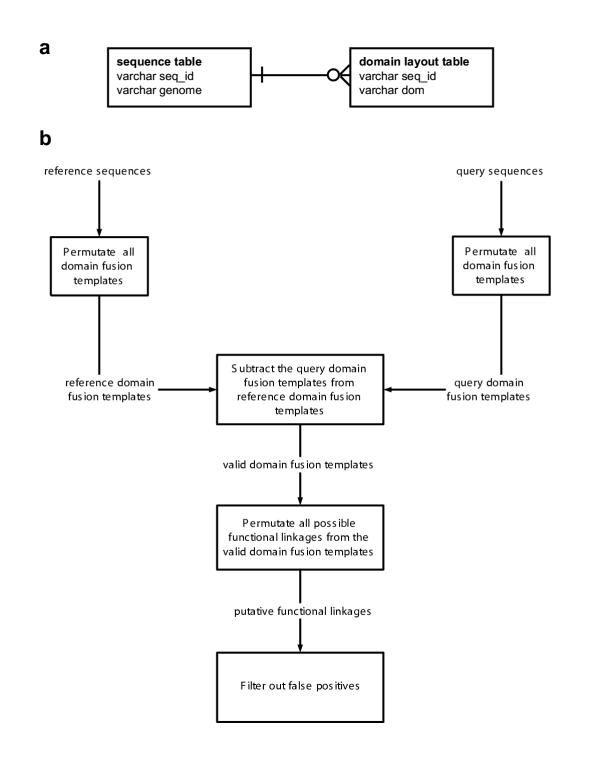
Figure 1.
(a) The Crow's Foot entity relationship diagram ofthe database architecture. (b) The flowchart of the method Protein sequences from SWISS-PROT+TrEMBL were divided into query sequences (belonging to the genome of interest) or reference sequences (everything else). All possible DFTs of the query or reference sequences were then permutated using a single SQL command. The valid DFTs were found by subtracting the query set from the reference set. Finally, using the valid DFTs, all functional linkages were permutated. The number of putative functional linkages is generally large, so it is necessary to filter out false positives.