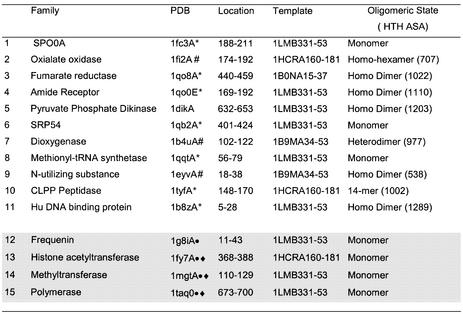
Table 3. There were 38 false positive proteins remaining when seven HTH templates were scanned against the PDB structures in CATH and a threshold rmsd value of <1.6 Å and ASA threshold of >990 Å2 applied.
This table gives details of the 15 structural families into which they were clustered using CATH. A hash symbol denotes structures in which the ASA of the HTH in the complete oligomer falls below the threshold of 990 Å2. * denotes structures in which one helix of the HTH identified is 3 or more residues longer than that in the HTH template. Circles denote those structures discussed in detail in Results and diamonds those structures proposed to have DNA-binding HTH motifs.