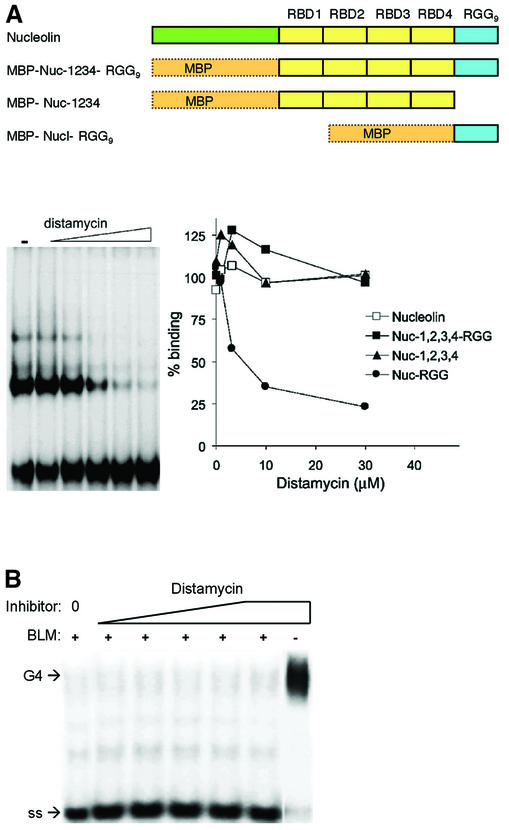
Figure 2.
Effect of distamycin on G4 DNA binding and unwinding. (A) (Top) Diagram of recombinant derivatives of nucleolin assayed as fusions to MBP. (Bottom) Effect of distamycin on G4 DNA binding by nucleolin and recombinant nucleolin derivatives. (Left) Distamycin inhibition of binding of Nuc-RGG to G4 DNA. [32P]G4 DNA was preincubated with 0, 0.12, 0.37, 1.11, 3.33 and 10 µM distamycin and then binding to 1 nM Nuc-RGG9 was assayed by gel mobility shift. (Right) Quantitation of binding assays carried out with purified murine nucleolin (Nucleolin, open square) and recombinant nucleolin carrying the RBDs and RGG9 (Nuc-1,2,3,4-RGG9, filled square), the nucleolin RBDs (Nuc-1,2,3,4, filled triangle) and the RGG9 domain of nucleolin (Nuc-RGG9, filled circle). (B) Effect of distamycin on BLM helicase G4 DNA unwinding activity. Unwinding assays were carried out in 0–125 µM distamycin. Control reactions contained distamycin, but no protein (right). Positions of G4 DNA substrate and single-stranded product (ss) are indicated.