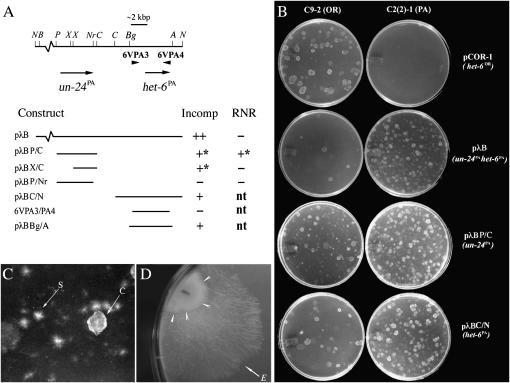
Figure 2.—
Incompatibility activity of constructs from the het-6 region. (A) Map at top gives selected restriction sites and locations of PCR primers used for making PA-haplotype constructs. Orientations of un-24PA and het-6PA are indicated by arrows. Restriction sites are A, ApaI; B, BamHI; Bg, BglII; C, ClaI; N, NotI; Nr, NarI; P, PstI; and X, XhoI. Incompatibility activity (Incomp) and ribonucleotide reductase catalytic function (RNR) for constructs are given below the map. Incompatibility activity was assayed by DNA transformations into OR [C9-2] and PA [C2(2)-1] spheroplasts and defined as strong (++), weak (+), weak with transformants having a star phenotype (+*; see text for details), or absent (−). Ribonucleotide reductase catalytic function is designated as “+” or “−” on the basis of whether or not the construct complements the un-24 temperature-sensitive mutation in C8c-164 spheroplasts. (B) Examples of DNA transformations that demonstrate strong (pCOR-1 and pλB) or weak (pλBP/C and pλBC/N) incompatibility activity. (C) Close up of star-like (“S,” self-incompatible) and cloud-like (“C,” wild type) morphologies observed when C9-2 (un-24OR) is transformed with un-24PA. (D) Fast-growing escape sector “E” evident after ∼7 days growth by a self-incompatible un-24PA transformant of C9-2 (arrowheads) following transfer from transformation plate to Vogel's medium.