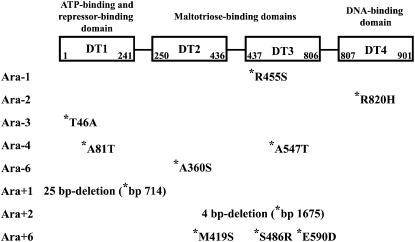
Figure 2.—
Schematic structure of the MalT protein and location of evolved mutant alleles. The protein is composed of four domains (Danot 2001; Richet et al. 2005): DT1 (residues 1–241), DT2 (residues 250–436), DT3 (residues 437–806), and DT4 (residues 807–901). DT1 binds ATP and contains surface determinants involved in the binding of repressor proteins, including Aes, MalK, and MalY. DT2 and DT3 are involved in maltotriose binding and represent putative multimerization domains. DT4 corresponds to the DNA-binding domain and presumably recruits RNA polymerase. The malT gene was sequenced in one clone isolated at 20,000 generations from each of the 12 populations designated Ara − 1–Ara − 6 and Ara + 1–Ara + 6. Amino-acid substitutions deduced from the mutations identified in the evolved clones are indicated using the one-letter code, and the residue number refers to its position in the MalT polypeptide. In both the Ara + 1 and Ara + 2 populations, small internal deletions were found in malT (their size and position relative to the malT translational start codon are shown).