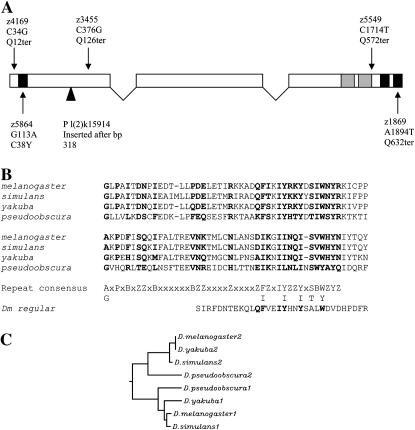
Figure 1.—
(A) Diagram of D. melanogaster tef transcript and mutations, with solid boxes indicating the positions of consensus sequences for C2H2 zinc-finger motifs and shaded boxes indicating a conserved repeat motif. For each mutation, the allele designation, alteration in DNA sequence, and predicted alteration in amino acid sequence are indicated. The insertion site of the P allele is indicated by a solid triangle. (B) Sequence alignment of a conserved tandem repeat, which resides in amino acid residues 478–573 in D. melanogaster (Z, hydrophilic; B, hydrophobic). The homologous repeat from the D. melanogaster regular gene is indicated below the consensus sequence. (C) ClustalW-generated dendrogram showing the degree of similarity between the two repeat domains identified in the carboxyl half of the predicted Tef protein. Branch lengths are proportional to degree of differences.