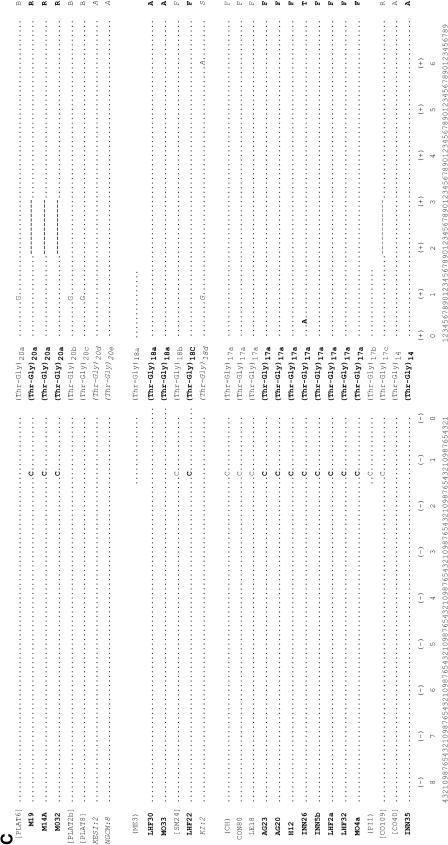
Figure 3.—
Thr-Gly flanking haplotypes from Europe, Australia, and Africa. DNA sequence of the regions flanking the uninterrupted Thr-Gly repeat of European and Australian (boldface type) and Kenyan (italic type) flies. Dots indicate conserved positions; dashes indicate deletions. The amino acid sequence is shown at the top and the only two amino acid substitutions (S/F and G/R) are indicated. The numbers at the bottom label each nucleotide position. Published sequences are reported in brackets. Note that the BX1, ME3, and PI1 sequences shown here are 3 bp longer, both in the 5′ and the 3′ regions flanking the main repeat, than in Costa et al. (1991); however, the sequences for the entire flanking regions were not available. Unless otherwise stated, the sequences described by Rosato et al. (1996) are shown in parentheses. The sequence prefixed with # is a personal communication (C. Aquadro). AG, Yeppoon (Queensland, Australia); AL, Albaida (Spain); BRFS, Bowen (Queensland, Australia); BX, Bordeaux (France); CH, Chieti (Italy) (Yu et al. 1987); CO, Cognac (France); CON, Conselve (Italy); CS, Canton-S (Jackson et al. 1986); DG, South Brisbane (New South Wales, Australia); GL, Hervey Bay (Queensland, Australia); H, Melbourne (Victoria, Australia); ICIPE, K, L, P, Nairobi (Kenya); INN, Cairns (Queensland, Australia); KESI, Kericho (Kenya); LE, Leiden (Netherlands); LEC, Lecce (Italy); LHF, Yeppoon (Queensland, Australia); M, Melbourne (Victoria, Australia); MA, Matuga (Kenya); ME, Merano (Italy); MO, Cairns (Queensland, Australia); NGMI and NGCM, Nguruman (Kenya); NW, North Wooton (United Kingdom); OR, Oregon-R (Citri et al., 1987); PI, Pietrastornina (Italy); PLAT, Platanistassa (Cyprus); SM, San Mateo (Spain).