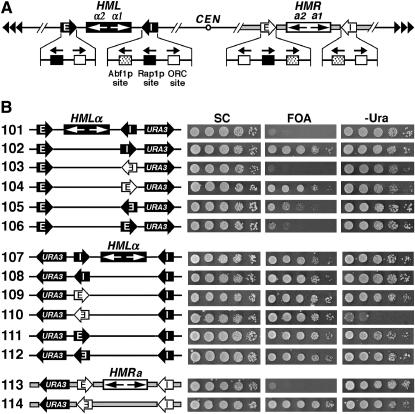
Figure 1.—
The apparent directionality of a silencer observed in a specific experiment depends on the genomic location of the silencer as well as the sensitivity of the silencing assay. (A) Schematics of the HML and HMR loci on chromosome III in S. cerevisiae. The HML-E and HML-I silencers are shown as solid arrows with white letters, and the HMR-E and HMR-I silencers are shown as open arrows with black letters. The direction of each silencer sequence is drawn as pointing toward the HMLα or HMRa genes, but is not necessarily the functional direction. The binding sites for Abf1p, Rap1p, and ORC in the silencers as well as their 5′ → 3′ directions are indicated. In HML-E, 16 bp centromere-proximal to the ORC site is a putative Sum1p-binding site whose deletion has been shown to reduce the activity of a weakened HML-E silencer (Irlbacher et al. 2005). The α1 and α2 genes at the HML locus and the a1 and a2 genes at HMR are also indicated. CEN, centromere. Tandem arrowheads, telomeric repeats. The regions spanning the HMR locus and flanking sequences are indicated by thick shaded lines. (B) Effects of the genomic contexts of HML-I and HML-E on the directionality of silencers. Left, the modified HML or HMR loci in strains 101–114 (see materials and methods for their construction). The silencers, HM genes, and the URA3 reporter gene are indicated. Cells of each strain were grown to late log phase and serial 10-fold dilutions were spotted on test plates and allowed to grow for 3 days. SC, synthetic complete medium. 5-FOA, SC supplemented with 1 mg/ml 5-fluoroorotic acid (5-FOA). −Ura, SC depleted of uracil. Growth phenotypes of each strain are shown on the right.