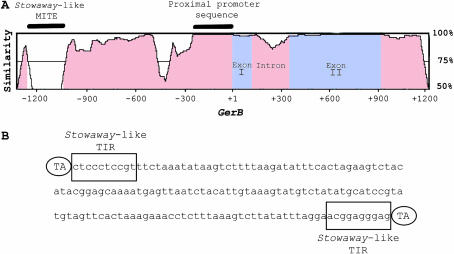
Figure 2.—
GerB and GerF are paralogous genes. (A) Barley cv. Morex GerB and GerF genomic sequence similarity is depicted in a VISTA plot. GerB sequence is plotted on the x-axis. Percentage similarity between GerF and GerB is plotted on the y-axis. The window size is 100 bp. Insertion of a Stowaway-like MITE in the GerB 5′ noncoding region (0% similarity) and 276 bp of highly conserved proximal promoter region (100% similarity) are denoted by solid bars. (B) Stowaway-like MITE sequence insertion (160 bp) present in GerB 5′ noncoding region. The first 10 bp of the terminal inverted repeats (TIRs) are conserved and match the maize Stowaway family of transposable elements consensus 5′-CTCCCTCCRT-3′ (boxed). The ovals depict the dinucleotide TA as target sequence duplications (TSDs).