TABLE 1.
Mapping results and mutations in nuclear-encoded genes with mitochondrial function that exhibit a G1–S block in the third larval instar with normal neuronal differentiation
| A. Mapping results | |||
|---|---|---|---|
 | |||
| Cytological position | Gene name | Mitochondrial process | Alleles |
| B. Mitochondrial genes identified by the glossy eye–BrdU screen | |||
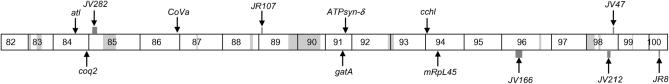
| 84E4 | CG10092 arginyl tRNA ligase (atl) | Protein biosynthesis–tRNA synthesis | JR70, JR77, JV83, JR101, JR203, JV219, JR247, JV250, JP782 |
| 84F11 | CG9613 coenzyme Q biosynthesis protein 2 (coq2) | Oxidative phosphorylation–complex III | JV259, JP768 |
| 91E4 | glutamyl-tRNA amidotransferase A (gatA) | Protein biosynthesis–tRNA synthesis | JR15, JV87, JR94, JR113, JR205 |
| 91F1 | ATP synthase subunit δ (ATPsyn-δ) | Oxidative phosphorylation–complex V of electron transport chain | JR92, JV115, JR238 |
| 94A1 | CG6022 cytochrome c heme lyase (cchl) | Oxidative phosphorylation–complex IV of electron transport chain | JV32, JV56, JV193, JR214; Exelixis c04553 (Thibault et al. 2004) |
| 94B6 | mitochondrial ribosomal protein large subunit 45 (mRpL45) | Protein biosynthesis–ribosome assembly | JV28 |
| Previously identified mitochondrial genes that cause the glossy-eye–defective BrdU incorporation phenotype | |||
| 23F3 | pdsw | Oxidative phosphorylation–complex I of electron transport chain | pdswk10101 (Spradling et al. 1999) |
| 35F11 | mitochondrial ribosomal protein large subunit 4 (mRpL4) | Protein biosynthesis–ribosome assembly | mRpL4k14608 (Spradling et al. 1999) |
| 61B3 | mitochondrial ribosomal protein large subunit 17 (mRpL17) | Protein biosynthesis–ribosome assembly | mRpL17KG06809 (Bloomington Drosophila Stock Center at Indiana University) |
| 86F9 | cytochrome c oxidase subunit Va (CoVa) | Oxidative phosphorylation–complex IV of electron transport chain | JP785 (Mandal et al. 2005) |
| Allele | Cytological position | No. of genes in region | Allele | Cytological position | No. of genes in region |
| Other mapping results | |||||
| JV282 | 85A6-B1 | 24 | JV212 | 98E1-E3 | 6 |
| JR107 | 89A5-A8 | 29 | JV47 | 98F5 | 9 |
| JV166 | 96A21-B4 | 34 | JR8 | 100B9 | 5 |
In A, dark shading indicates complementation groups that have mapped to a deficiency and light shading indicates gaps within the available deficiency kit. The candidate regions and potential mitochondrial genes for complementation groups that have not been cloned are also indicated.
