TABLE 2.
Codon model parameter estimates for AP1/FUL paralogs
| Model | p | ℓ | Estimates of parameters | Positively selected sites |
|---|---|---|---|---|
| M0: one ratio | 1 | −3714.44 |
 = 0.1181 = 0.1181 |
None |
| Branch-specific models | ||||
| Two ratios | 2 | −3714.27 |
 0 = 0.1167, 0 = 0.1167, 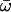 1 = 0.1525 1 = 0.1525 |
None |
| Three ratios | 3 | −3714.22 |
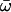 0 = 0.1223, 0 = 0.1223, 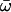 1 = 0.1527 1 = 0.1527  2 = 0.1147 2 = 0.1147 |
None |
| Site-specific models | ||||
| M1a: near neutral | 2 | −3635.68 | p0 = 0.87366, (p1 = 0.12634) | Not allowed |
| M2a: selection | 4 | −3635.68 |
p0 = 0.87368, p1 = 0.12632 (p2 = 0.00000),  2 = ∞ 2 = ∞ |
None |
| M3: discrete (K = 3) | 3 | −3614.81 |
p0 = 0.52591, (p1 = 0.39457), p2 = 0.07953, 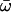 0 = 0.01531 0 = 0.01531 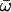 1 = 0.18259, 1 = 0.18259, 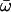 2 = 0.89122 2 = 0.89122 |
None |
| M7: β | 2 | −3618.86 | p = 0.32839, q = 1.74796 | Not allowed |
| M8: β and ω | 4 | −3619.31 |
p0 = 0.99707, p = 0.32866 q = 1.75003, (p1 = 0.00293) 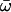 = 118.19 = 118.19
|
167V (P = 0.86) 207T (P = 0.60) 208T (P = 0.73) 215L (P = 0.68) |
| Branch-site models | ||||
| Model A: FUL1 (= null Model A) | 4 | −3635.68 |
p0 = 0.87366, p1 = 0.12634, (p2a + p2b = 0.000) 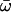 0 = 0.0729 0 = 0.0729  2a = 1.00000, 2a = 1.00000, 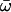 2b = 1.00000 2b = 1.00000 |
None |
| Model B: FUL1 (= null Model B) | 5 | −3626.84 |
p0 = 0.80202, p1 = 0.19798, (p2 + p3 = 0.00),  0 = 0.05027 0 = 0.05027 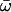 1 = 0.52984, 1 = 0.52984, 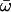 2 = ∞ 2 = ∞ |
None |
| Model A: FUL2 (= null Model A) | 4 | −3635.19 |
p0 = 0.77157, p1 = 0.11071 (p2a + p2b = 0.11773) 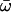 0 = 0.0714, 0 = 0.0714,  2a = 1.00000 2a = 1.00000  2b = 1.00000 2b = 1.00000 |
45L (P = 0.69) 50T (P = 0.62) 145Q (P = 0.61) 206D (P = 0.51) |
| Model B: FUL2 (= null Model B) | 5 | −3626.84 |
p0 = 0.80202, p1 = 0.19798, (p2 + p3 = 0.00),  0 = 0.05027 0 = 0.05027  1 = 0.52984, 1 = 0.52984, 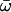 2 = ∞ 2 = ∞ |
Parameter estimates that are underlined indicate positive selection; those in parentheses are not free parameters (p). Putative sites under positive selection are based on the coding region of rice OsMADS14. Estimates of κ ranged between 1.74 and 1.9. ℓ, log likelihood; p, number of parameters; q, free parameter.
