Figure 2.
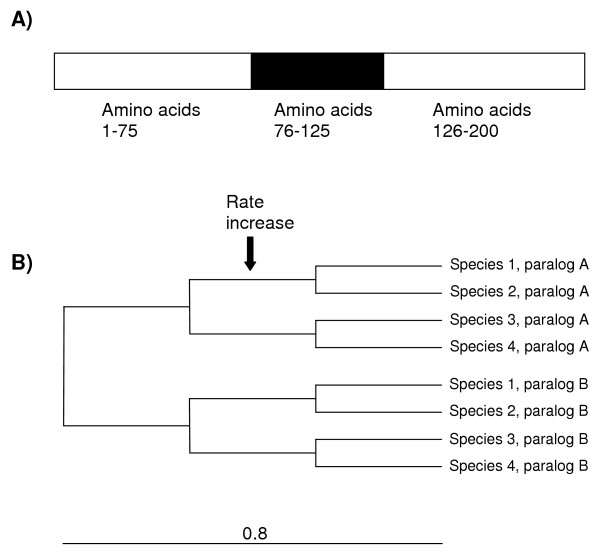
A) Structure of the simulated proteins. Total length was 200 amino acids. The region in which different evolutionary rates were tested correspond to positions 76–125. B) Topology of the tree in which the simulations were performed. Total branch length is 0.8 and the length of all inner branches is 0.8/3 = 0.267. The default average substitution rate was set to μ = 0.5. This rate was increased to μ = 1, μ = 2.5 or μ = 5 in the 76–125 region to test for the effect of local rate increase on UVPAR results. The α parameter of the gamma distribution of rate heterogeneity among sites was varied between 0.2 and 3 (see text and Table 1). A total of 100 simulations were performed for each combination of μ and α.
