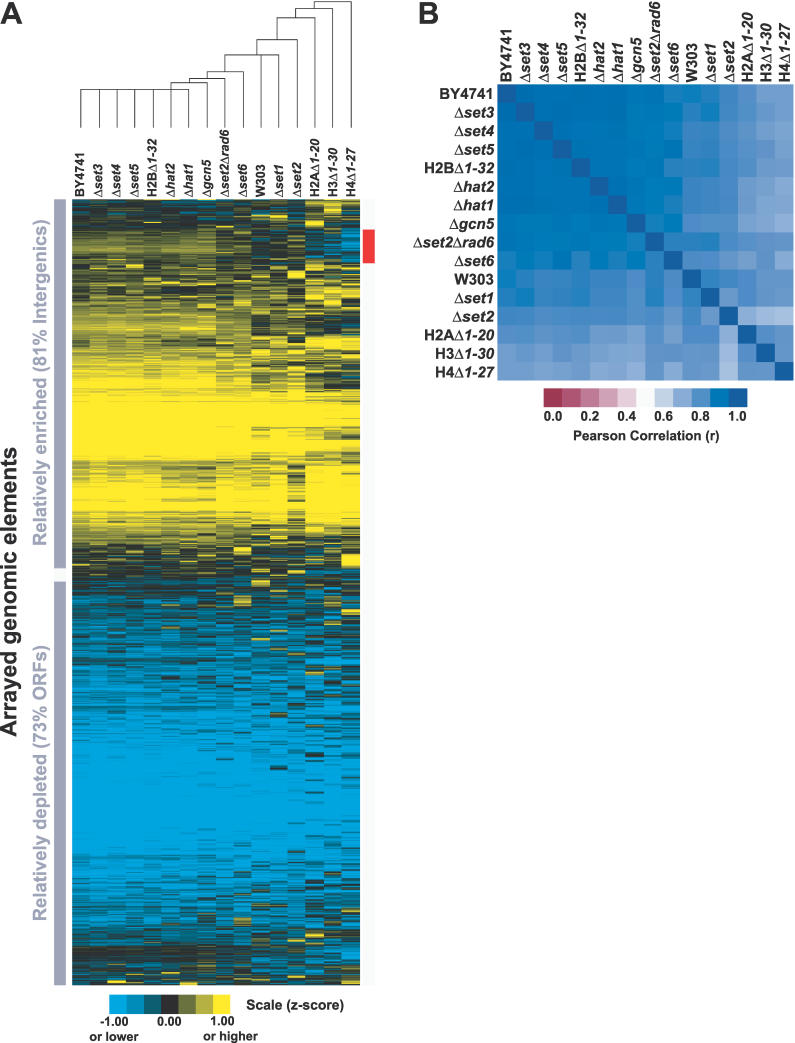
Figure 3. FAIRE Is Unaffected by Mutations in Genes Required for Specific Histone Modifications.
(A) Data are from FAIRE performed on 15 mutant strains and two wild-type strains. For each strain, normalized median log2 ratios from individual arrays were z-score transformed and averaged with biological replicates (Material and Methods). Hierarchical clustering by gene and by array was performed on all arrayed elements using Cluster [57]. The red bar indicates the 493 loci that segregate most differently in the histone tail deletion mutants. Nearly all of them (459/493, 93%) are intergenic regions. Of the 34 ORFs that differentially segregate, 12 are annotated as “dubious.” Analysis revealed no striking difference between the 493 loci and the rest of the genome with respect to GC content, and these loci are not enriched for subtelomeric or telomeric fragments. The expression level and rate of transcription of genes downstream of the intergenic loci were slightly lower than the genomic average.
(B) The Pearson correlations of the FAIRE data from the 17 different mutant and wild-type strains.