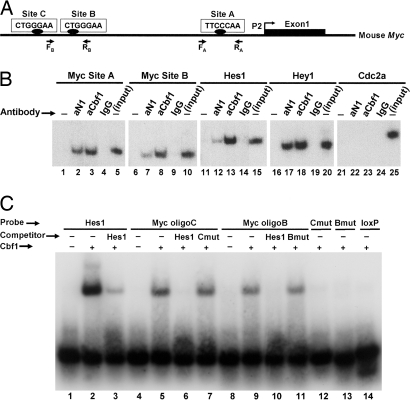
Fig. 4.
Analysis of Cbf1-binding sites in the mouse Myc promoter. (A) Shown is a diagram of the mouse Myc gene 5′ flanking region upstream from the first exon transcribed by activation of the most frequently used promoter P2. The positions of putative Cbf1-binding sites (sites A–C) and of the PCR primers used in ChIP experiments are indicated. (B) ChIP assays. In the examples shown, chromatin from a nonregressing N1IC-induced tumor was immunoprecipitated with antibodies against Notch1 (aN1) or Cbf1 (aCbf1) or with control rabbit IgG and analyzed by PCR using primers specific for the indicated promoters. The Myc promoter was analyzed for sites A and B. Hes1 and Hey1 were examined as positive controls; Cdc2a served as a negative control. “Input” corresponds to products generated by PCR (still in exponential phase; 25 cycles) by using DNA extracted from nonimmunoprecipitated chromatin (10% of the amount used in experimental samples) as a template. −, no antibody was added to the reaction mixture. (C) EMSA was performed by using a recombinant Cbf1 protein fragment and the indicated labeled oligonucleotide probes. Myc oligoB and oligoC span, respectively, the corresponding Cbf1 sites shown in A. An oligonucleotide representing two known Cbf1-binding sites of the Hes1 promoter was used as a positive control. (We attribute the shift of two bands to the presence of these two sites.) Mutated versions of Myc oligonucleotides B (Bmut) and C (Cmut) and an unrelated loxP oligonucleotide served as negative controls. In the competition assays, unlabeled Hes1, Bmut, and Cmut oligonucleotides were added to the binding mixture at a 50-fold molar excess.