Figure 6.
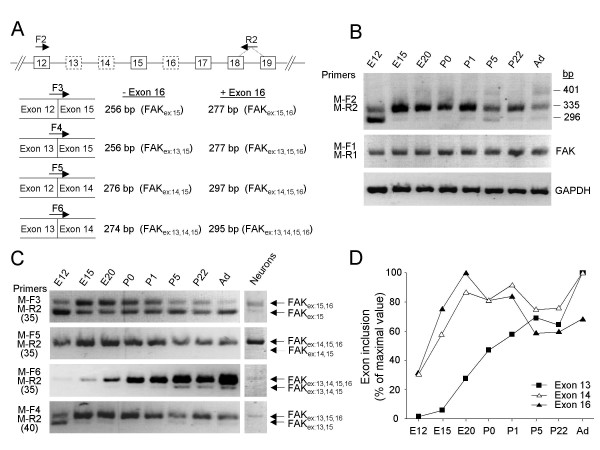
Alternative splicing of FAK exons 13, 14 and 16 in mouse brain during development. (A) RT-PCR strategy. Top: Genomic organization of the FAK gene (exons 12–19); alternatively spliced exons are boxed with dotted lines. The positions of the primers are indicated. PCR with the flanking forward (F2) and reverse (R2) primers amplified simultaneously all exon combinations. The selective targeting of FAK alternative transcripts was achieved with specific forward primers (F3, F4, F5, F6) coupled to the R2 primer. Each forward primer targeted transcripts with only one combination of exons 12, 13, 14 and 15; the size of the PCR product(s) distinguishes the absence or presence of exon 16 in each transcript. For each species similar primers were used (termed according to the species and the position, see additional file 8), taking into account sequence variations. (B) RT-PCR analysis of alternative splicing of exons 13, 14 and 16 during mouse brain development using flanking primers. The position of molecular markers 296 bp, 335 bp and 401 bp correspond to the size of the expected PCR products for FAKex:15, FAKex:14,15,16 and FAKex:13,15,16 respectively. The total levels of FAK transcripts were estimated by RT-PCR with primers (M-F1 and M-R1) located in constitutive exons. Amplification of GAPDH transcripts was used as a control. (C) RT-PCR monitoring the developmental expression of alternatively spliced FAK transcripts (left panel) and their expression in E16 cortical neurons (right panel) are shown. The number of PCR cycles used is indicated between parenthesis. (D) Pattern of inclusion of FAK alternative exons 13, 14 and 16 during development. Quantification of PCR products was performed as described in Methods. The inclusion of exons 13, 14 or 16 was estimated at each developmental stage by adding the normalized values of all the transcripts containing the corresponding exon and expressed as percent of the maximum.
